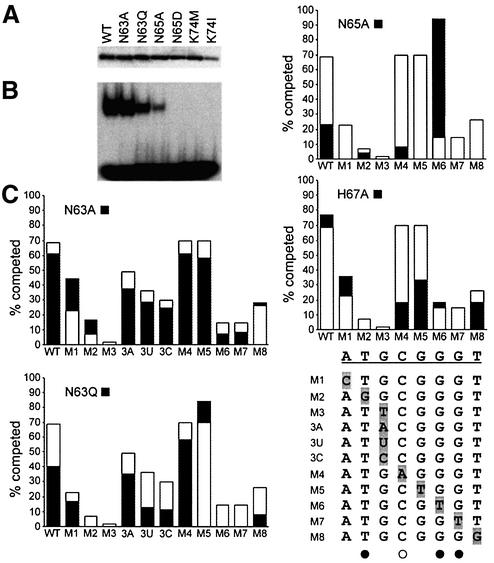
Fig. 5. DNA-binding properties of mutant GCM domains. (A) Expres sion of T7-epitope tagged wild-type (WT) and mutant (N63A, N63Q, N65A, N65D, K74M, K74I) GCM domains was verified by western blot of nuclear extracts from transfected COS cells with a monoclonal antibody against the tag. (B) Electrophoretic mobility shift assay with the consensus GCM binding site as probe and extracts from transfected COS cells expressing the wild-type and mutant GCM domains. Equal amounts of each GCM domain were used. (C) Comparative DNA- binding analysis of wild-type GCMa and GCM protein mutants by competition analyses. Electrophoretic mobility shift assays were performed with the consensus GCM binding site as probe and extracts expressing the wild-type and mutant GCM protein in the absence and presence of increasing amounts of competitor (5-, 10-, 25-, 50- and 100-fold molar excess). Oligonucleotides containing the consensus GCM binding site (WT) and its variants (M1–M8, 3A, 3U, 3C) were used as competitors. Conditions were such that in the absence of competitor, 20–30% of the radioactively labeled probe was in complex with the GCM domain. The competitor-dependent reduction of probe in the complex was determined by phosphoimager analysis. The graph summarizes the relative level of competition obtained with a 10-fold excess of each competitor (WT, M1–M8, 3A, 3U, 3C) for wild-type GCMa (open bars) and GCM mutants (black bars). WT and mutant target sites (M1–M8, 3A, 3U, 3C) are listed. Directly and indirectly contacted bases as observed in the crystal structure are marked with filled and open circles, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.