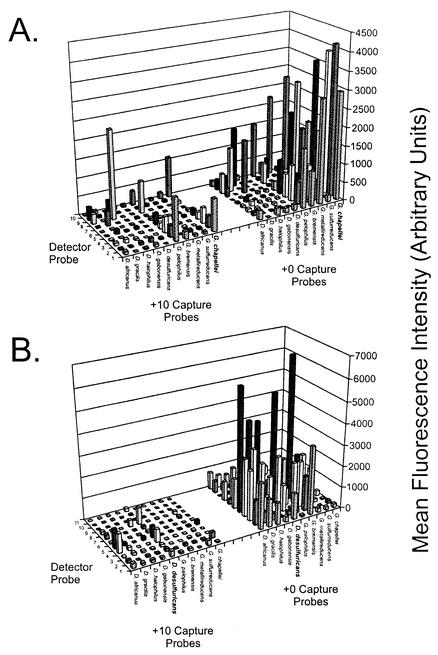
FIG. 3.
Effect of nucleotide mismatches between the detector probe and target rRNA. Detector probe sequences and perfect matches are listed in Table 1. Probes1 to 3 are specific for Geobacter species; probes 4 to 8 are specific for Desulfovibrio species; probe 9 is specific for S. putrefaciens; and probe 10 is identical to probe 1 and is specific for Geobacter species, except that it is shortened by 4 nt to achieve a +14 hybridization condition. Signals represent the average hybridization intensity from at least three replicate arrays (across two slide lots), with each array containing three replicate spots. Hybridizations were performed as described in Materials and Methods with 2 μg of fragmented G. chapellei target RNA (A) or fragmented D. desulfuricans target RNA (B). Similar results were obtained with intact RNA targets (not shown). The graphing software did not permit error bars to be plotted in the three-dimensional graphing format, and so they are not shown.