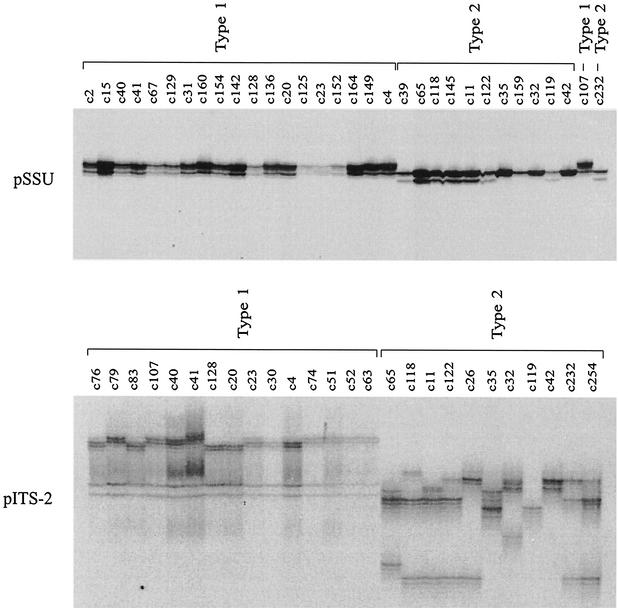
FIG. 5.
Representative DPGE analysis of sequence variation in the two loci, pSSU (top) and pITS-2 (bottom), within and among C. parvum samples from humans. Subsets from the 184 samples scanned at both loci by SSCP, and representing the entire spectrum of sequence variation detected have been included. For pSSU, the first 19 samples (c2 to c4) were identified as type 1, and the following 11 samples (c39 to c42) were identified as type 2 (Fig. 1). Samples c107 and c232 from humans from the United Kingdom were included as type 1 and type 2 reference controls, respectively. For pITS-2, the first 4 samples (c76 to c107) from a nursery outbreak in Middlesex (United Kingdom) and the next 11 samples (c40 to c63) from humans following recent foreign travel were identified as type 1. Another nine samples (c65 to c42) from patients following overseas travel and two samples (c232 and c254) from a waterborne outbreak at Clitheroe (United Kingdom) were identified as type 2. Significant pITS-2 profile variation was detected within both types 1 and 2. The level of sequence variation displayed within and among samples representing type 2 was greater than that for type 1, in accordance with SSCP analysis (Fig. 1).