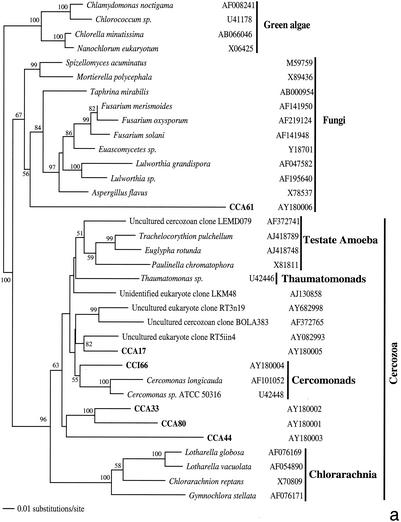
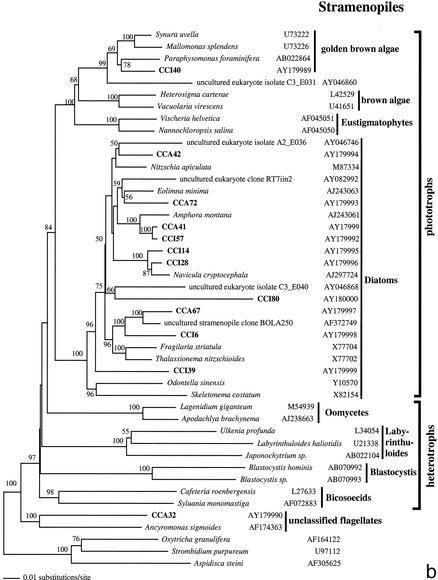
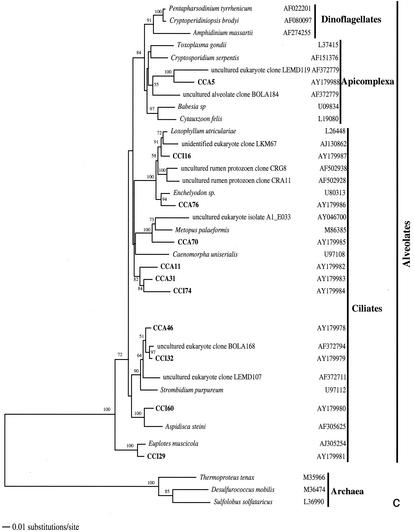
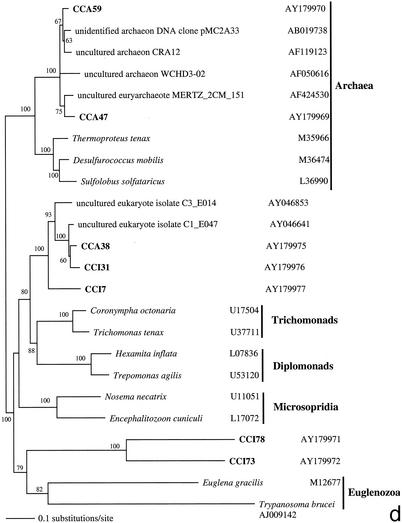
FIG. 2.
Phylogenetic trees for nearly complete SSU rRNA sequences of environmental clones in the Great Sippewisset salt marsh, Cape Cod, Mass., and the most closely related cultured organisms and environmental 18S rRNA signatures. Environmental clones are indicated by boldface type, and each clone is designated by the library designation (CCI for water-sediment interface; CCA for subsurface sediment [10 cm]) followed by a number. The tree topologies were obtained by a heuristic search with stepwise addition of taxa (TBR branch swapping) under a Tamura Nei minimum evolutionary distance model (trees a, b, and c) and maximum-likelihood (tree d) trees, with gapped and ambiguously aligned positions excluded. Distance bootstrap values over 50% from an analysis of 1,000 bootstrap replicates are given at the respective nodes. Sequences are followed by GenBank accession numbers.