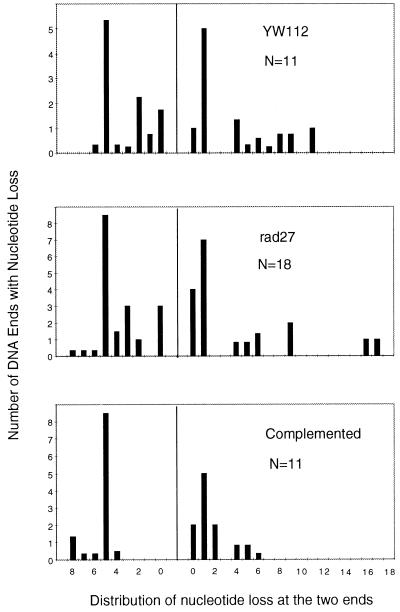
Figure 2.
Comparison of nucleotide loss from the miscellaneous category of products in YW112, rad27, and the complemented strain. A histogram is shown for double-strand break junctional sequences, showing the number of ends along the y axis and the number of nucleotides lost on the x axis. N is the total number of ends scored for each histogram. When the precise number of nucleotides lost from an end was ambiguous because of microhomology use at the junction, the nucleotide loss from each end was calculated as half of the nucleotides lost from the junction; then, the loss was evenly distributed among all potential positions within each end.