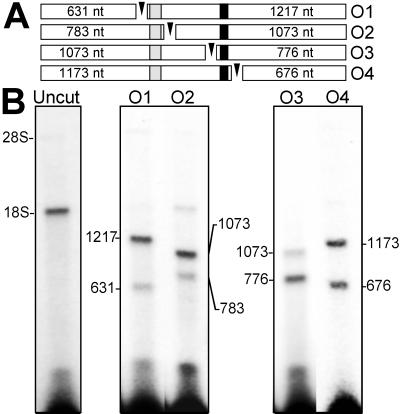
Figure 3.
Mapping the binding site of the GTX RNA probes by RNase H digestion of the rRNA (A). The open bars represent the 18S rRNA fragments after RNase H digestion, the sizes of which are indicated within each bar. The shaded and solid segments indicate the positions of GTX complementarity at nucleotides 701–741 and 1104–1136, respectively. The oligonucleotides (O1–O4) used to direct digestion of rRNA by RNase H are represented as arrowheads. O1 is complementary to the 18S rRNA at nucleotides 632–652, O2 is complementary at nucleotides 784–796, O3 is complementary at nucleotides 1074–1093, and O4 is complementary at nucleotides 1174–1193. (B) Results of the RNase H digestions. After cross-linking the GTX-1 or GTX-2 probe to dissociated ribosomes, the RNAs were purified away from protein and annealed to the O1 or O2 oligonucleotides (for the GTX-1 probe) or to the O3 or O4 oligonucleotides (for the GTX-2 probe), and the rRNA at these positions was digested with RNase H. The positions of the rRNAs were determined by ethidium bromide staining of the agarose gels.