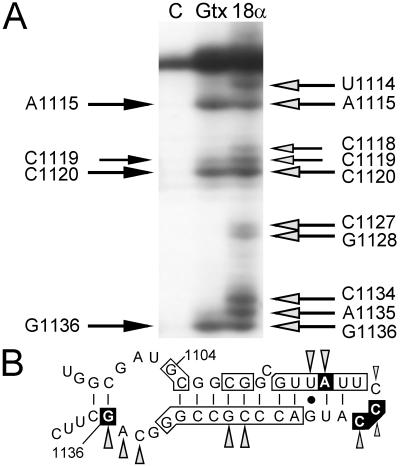
Figure 5.
Reverse transcription analysis of one region of 18S rRNA after cross-linking of RNA probes to ribosomes. (A) Lanes contain primer extension products from control ribosomes without probe (lane C), from ribosomes cross-linked to the GTX-2 probe (lane Gtx), and from ribosomes cross-linked to the 18Sα(1108) probe (lane 18α). The black arrows to the left of the autoradiogram indicate the nucleotide positions where reverse transcriptase stopped when ribosomes were cross-linked to the GTX-2 probe. Similarly, the shaded arrows to the right of the gel indicate stop sites in the presence of the 18Sα(1108) probe. All of these sites have been seen in at least four experiments. (B) Schematic representation of 18S rRNA at nucleotides 1097–1140 indicating complementarity to Gtx mRNA and summary of toeprinting results. The secondary structure of this segment of the 18S rRNA is based on Holmberg et al. (30). Vertical lines indicate base pairing and the solid dot represents G⋅U base pairing. Complementarity between the Gtx mRNA and the 18S rRNA is indicated by the boxed nucleotides. The nucleotide positions at which reverse transcriptase stopped in the presence of the GTX-2 probe are boxed in black. Stop sites in the presence of the 18Sα(1108) probe are indicated as shaded arrowheads.