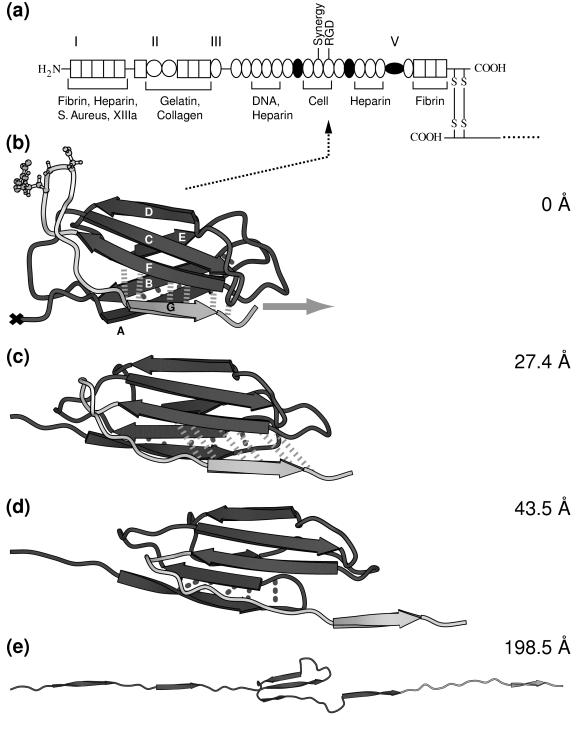
Figure 1.
(a) Modular structure of the fibronectin family. Fibronectin consists of two monomeric strands linked by two disulfide bridges (S—S). Each monomer contains three types of modules, types I, II, and III, and/or variable modules V. Modules that are either inserted or missing in various spliced forms of fibronectin are shown as filled circles. The cell adhesion site and other recognition sites are indicated. (b–e) Forced unfolding sequence of FnIII10 for various extensions of the module. A different gray scale is used to highlight the RGD motif and strand G. Hydrogen bond pairs that are broken in clusters during the unfolding burst of the module are displayed by dotted (AB bond pairs) and dashed (FG bond pairs) lines. The ball-and-stick representation indicates the position of the RGD cell binding region along the FG loop of FnIII10. Water molecules that were included in the SMD simulation are omitted for clarity. The equilibrated structure of FnIII10 is shown in b with its fixed terminal end (×) and the applied external pulling force (gray arrow). Partial distortion of the module is evident in c with the hydrogen bonds between strand F and G being stretched. Finally, in d, strand G clearly is separated from β-sheet FCD with the hydrogen bonds between F and G broken apart. Only strands C, D, and E remain in e to form a structural unit whereas the other strands form a line along the pulling direction. Further pulling on the module leads to total disintegration of the module with all strands aligned. This figure was created with molscript (37).