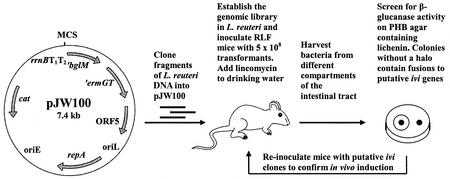
FIG. 1.
Selection of genes exhibiting elevated expression in the gastrointestinal tract of mice. Genes displaying elevated levels of expression in the intestinal tract were selected on the basis of their ability to drive the expression of a gene that is essential for colonization. Lactobacilli cannot become established in the guts of RLF mice treated with lincomycin administered in the drinking water. Colonization can occur only if an active promoter is inserted upstream of the promoterless ermGT gene. Recovery of strains from the gut after selection results in the detection of promoters that are either constitutive or ivi promoters. To distinguish between these two types of promoters, a promoterless marker gene (′bglM) is transcriptionally fused to ′ermGT, which enables screening for promoter activity in vitro. BglM− strains (no halo after agar plates containing lichenin are flooded with Congo red) or strains with low β-glucanase activity contain fusions to promoters activated in response to the gastrointestinal tract. ′ermGT, erythromycin resistance determinant; ′bglM, β-glucanase gene; cat, chloramphenicol resistance determinant; rrnBT1T2, E. coli rho-independent transcription terminators; repA, Lactobacillus replication gene; oriL, Lactobacillus origin of replication; oriE, E. coli origin of replication (pUC29); ORF5, ORF originating from pGT232.