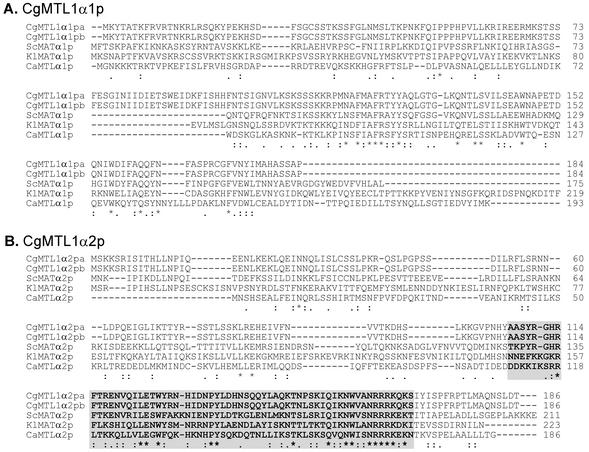
FIG. 4.
Sequence comparison of MTL1α1p and MTL1α2p of Candida glabrata strains PB921 (CgMTL1α1pa and CgMTL1α2pa) and 1480.47 (CgMTL1α1pb and CgMTL1α2pb) with MATα1ps or MTLα2ps and MATα2ps or MTLα2ps of other yeast species. (A) Aligned sequences of C. glabrata MTLα1pa and MTLα1pb with MATα1p of S. cerevisiae (Sc), MATα1p of K. lactis (Kl), and MTLa1p of C. albicans (Ca). (B) Aligned sequences of C. glabrata MTL1α2pa with MATα2p of S. cerevisiae, MATα2p of K. lactis, and MTL1α2p of C. albicans. The shaded sequences in panel B and the method used to indicate conserved amino acid sequences are described in the legend to Fig. 3. The accession numbers for the sequences other than those of C. glabrata are as follows: ScMATα1p, AAA34763.1; KlMATα1p, AAG21092.1 (note that the carboxy-terminal 42 amino acids of this K. lactis sequence are not shown); CaMATα1p, AAD51411.1; ScMATα2p, AAA34762.1; KlMATα2p, AAG21091.1; and CaMATα2p, AAD51408.1. The accession numbers for C. glabrata MTL1αa, MTL1αb, and MTL3α are AY191462, AY191463, and AY207368, respectively.