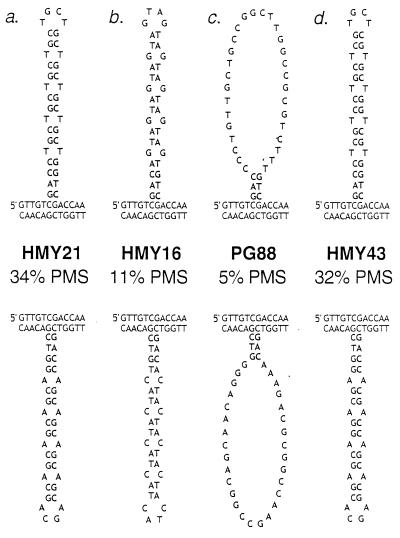
Figure 3.
Expected secondary structures in DNA loops containing various insertions within HIS4. As discussed in the text, heteroduplex formation between a his4 gene with a mutant insertion and a wild-type HIS4 gene will produce a DNA loop. The upper strand in each duplex is the nontranscribed strand. (Upper) The loops result from a heteroduplex in which the nontranscribed strand is mutant and the transcribed strand is wild type. (Lower) The nontranscribed strand is wild type and the transcribed strand is mutant. The inserted sequence is shown in bold. Strain names and the percentages of the unselected tetrads representing PMS tetrads are shown in the middle. A high percentage of PMS reflects inefficient repair of the DNA loop. The relevant genotypes are: (a) HMY21 (heterozygous for his4-CTG10); (b) HMY16 (heterozygous for his4-AGT10); (c) PG88 (heterozygous for his4-R), his4-R represents an insertion of 36 bp with a randomized DNA sequence that should be incapable of forming stable secondary structures, and (d) HMY43 (heterozygous for his4-Pal), his4-Pal is a 36-bp, nontriplet repeat sequence that should have similar base-pairing properties to his4-CTG10.