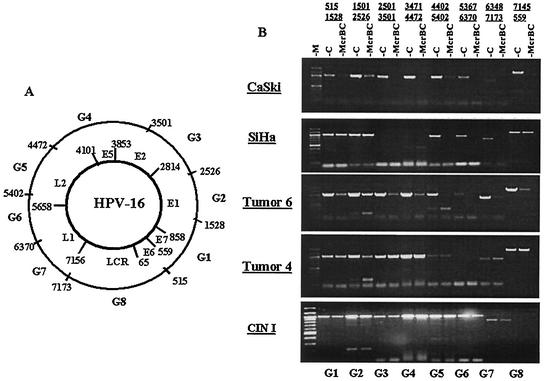
FIG. 2.
McrBC cleavage of segments of the HPV-16 genomes in SiHa and CaSki cells and three clinical samples detects patterns of hyper- and hypomethylation. (A) Genomic map of HPV-16 (7,906 bp), with genes E6, E7, E1, E2, E5, L2, and L1, the LCR, and the relative locations of amplicons G1 to G8 indicated. (B) Cleavage of amplicons G1 to G8 by McrBC (right lane of each pair of samples) indicates methylation, compared with uncleaved controls (left lane of each pair). The cleavage pattern indicates hypermethylation throughout most HPV-16 genomes in DNA from CaSki cells and tumor 6, hypermethylation of the late genes in the single HPV-16 genome of SiHa cells, and hypo- or no methylation in tumor 4 and a CIN I lesion.