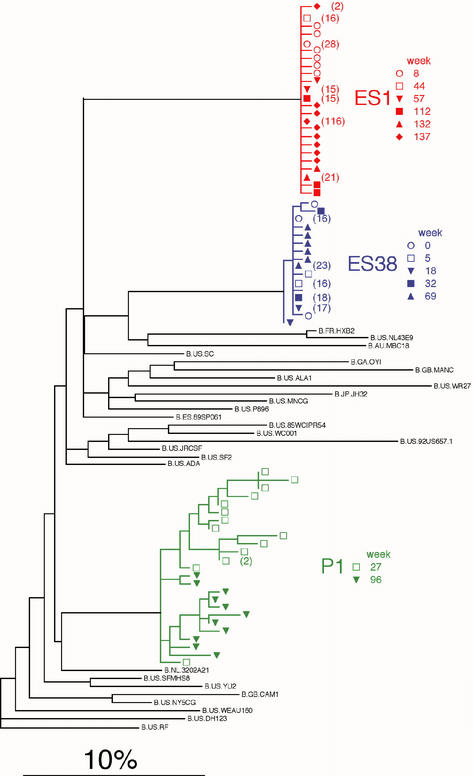
FIG. 4.
Phylogenetic analysis of HIV-1 env V3-C3 sequences of ES1 and ES38. An MLE phylogram (lnL = −269.2349) was constructed by using PAUP* illustrating the inferred evolutionary relationships of sequences obtained from ES1, ES38, and the HIV-1-infected sexual partner of ES1 (P1) and from the Los Alamos Retrovirus Database (24) and a database of local sequences by using FASTA (26). The model of evolution (GTR+G) was selected by using the AIC (1) in Modeltest version 3.06 (38). The parameters of this model were as follows: equilibrium nucleotide frequencies, fA = 0.4632, fC = 0.1299, fG = 0.1971, and fT = 0. 2098; shape parameter (α) of the 71 distributions reflecting the site-to-site rate variability of variable sites, α = 0.7535; R matrix values, RA<->C = 2.531, RA<->G = 2.962, RA<->T = 0.655, RC<->G = 1.571, RC<->T = 4.728, and RG<->T = 1. The scale bar represents the 10% MLE sequence distance. Multiple identical sequences from ES1, ES38, and P1 were represented by a single sequence in the phylogenetic reconstruction analyses; the numbers of sequences represented by a single node are shown in parentheses.