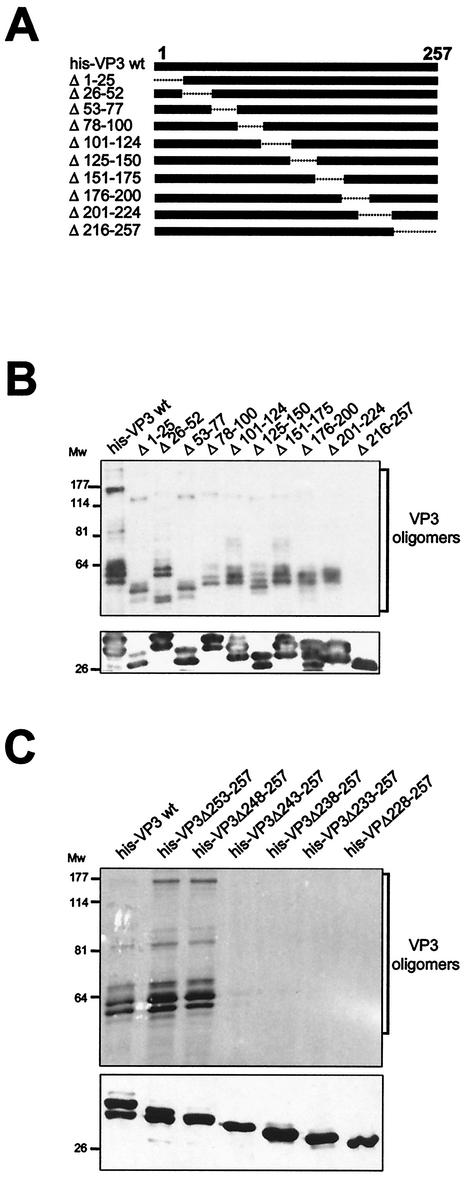
FIG. 7.
VP3 oligomerization domain mapping. (A) The diagram shows the set of VP3 deletion mutants used for mapping the VP3 oligomerization domain. Deleted regions are depicted as dashed lines. The name of each mutant refers to the boundaries of the deleted region. (B) Detection of VP3 oligomers. His-VP3 proteins were affinity purified, subjected to SDS-PAGE and Western blot analysis with rabbit anti-VP3 serum, followed by addition of horseradish peroxidase-conjugated goat anti-rat immunoglobulin. The signal was detected by enhanced chemiluminescence. Blots were excised, and the upper half was overdeveloped to facilitate the detection of oligomers. The lowerhalf, containing His-VP3 monomers, was developed with a shorter exposure. The positions of molecular mass markers (in kilodaltons) are indicated. (C) Detection of VP3 oligomers produced by C-terminal His-VP3 deletion mutant proteins. Proteins were affinity purified, subjected to SDS-PAGE and Western blot analysis with rabbit anti-VP3 serum, followed by addition of horseradish peroxidase-conjugated goat anti-rat immunoglobulin. The signal was detected by enhanced chemiluminescence. Blots were excised, and the upper half was overdeveloped to facilitate the detection of oligomers. The lower half of the blot, containing His-VP3 monomers, was developed with a shorter exposure time. The positions of molecular mass markers (in kilodaltons) are indicated.