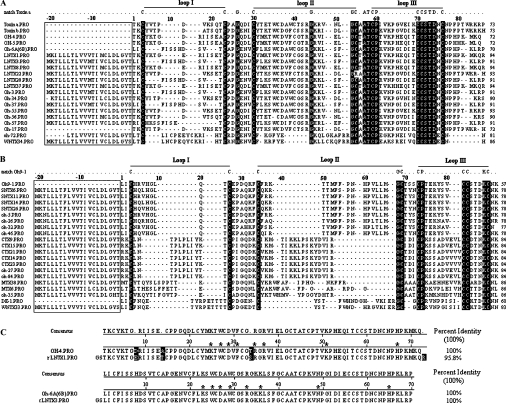
Figure 2. Multiple sequence alignments of deduced amino acids with those of toxins from O. hannah.
Natural peptides for which sequences have already been published were Toxin a (Swiss-Prot. number P01387), Toxin b (P01386), OH-4 (P80516), OH-5 (P80965), Oh-6A/6B (P82662), Oh9-1 (P83302) and DE-1 (P01412). The sequences of Oh-3, -5, -17, -26, -27, -32, -34, -35, -37, -46, -55, -56, -57 and -84 were deduced from their corresponding cDNAs with GenBank® accession numbers from AY596925 to AY596940 [23]. Signal peptides are boxed. Amino acid residues that matched the template residues in the first line of each group reversed-out (i.e. white-on-black) in (A) and (B), whereas residues reversed-out in (C) are unmatched ones. The last number in each row indicates the numbers of residues in each toxin. (A) Group A toxins having ten conserved cysteine residues (five disulphide bridges). (B) Group B toxins having eight conserved cysteine residues (four disulphide bridges). (C) Alignments of rLNTX1 and rLNTX3 to their corresponding natural proteins. Functional residues at homologous sites are indicated with asterisks (*).