Abstract
Adipose tissue is a major endocrine organ that exerts a profound influence on whole-body homoeostasis. Two types of adipose tissue exist in mammals: WAT (white adipose tissue) and BAT (brown adipose tissue). WAT stores energy and is the largest energy reserve in mammals, whereas BAT, expressing UCP1 (uncoupling protein 1), can dissipate energy through adaptive thermogenesis. In rodents, ample evidence supports BAT as an organ counteracting obesity, whereas less is known about the presence and significance of BAT in humans. Despite the different functions of white and brown adipocytes, knowledge of factors differentially influencing the formation of white and brown fat cells is sparse. Here we summarize recent progress in the molecular understanding of white versus brown adipocyte differentiation, including novel insights into transcriptional and signal transduction pathways. Since expression of UCP1 is the hallmark of BAT and a key factor determining energy expenditure, we also review conditions associated with enhanced energy expenditure and UCP1 expression in WAT that may provide information on processes involved in brown adipocyte differentiation.
Keywords: brown adipose tissue (BAT), peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α), signalling, transcription, white adipose tissue (WAT)
Abbreviations: AR, adrenergic receptor; BAT, brown adipose tissue; CBP, CREB-binding protein; C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; Cidea, cell death-inducing DFF45-like effector A; CPT-1b, carnitine palmitoyltransferase 1b; CREB, cAMP response element-binding protein; Dio2, type 2 iodothyronine deiodinase; 4E-BP1, 4E-binding protein 1; eNOS, epithelial NO synthase; ERR, oestrogen-related receptor; ES, embryonic stem; FABP, fatty acid-binding protein; Foxc2, forkhead box C2; GABP, GA-binding protein; IGF, insulin-like growth factor; IGF-1R, IGF-1 receptor; IRS, insulin receptor substrate; LRH-1, liver receptor homologue 1; LXR, liver X receptor; MAPK, mitogen-activated protein kinase; MEF, mouse embryo fibroblast; mTOR, mammalian target of rapamycin; NCoR, nuclear receptor co-repressor; NO, nitric oxide; NRF, nuclear respiratory factor; p/CIP, p300/CBP/co-integrator-associated protein; PGC-1, PPARγ co-activator 1; PKA, cAMP-dependent protein kinase; PKB, protein kinase B; PPAR, peroxisome proliferator-activated receptor; pRB, retinoblastoma protein; PRC, PGC-1-related co-activator; PRMT1, protein arginine methyltransferase 1; Rb, retinoblastoma gene; RIP140, receptor interacting protein 140; RXR, retinoid X receptor; SHP, small heterodimer partner; SIRT, sirtuin; S6K, S6 kinase; SNS, sympathetic nervous system; SPPARM, selective PPARγ modulator; SRC-1, steroid receptor co-activator 1; SREBP-1, sterol regulatory element-binding protein 1; SV, stromal-vascular; SV40, simian virus 40; T3, 3,5,3′-tri-iodothyronine; T4, thyroxine; TAg, large T antigen; Tfam, mitochondrial transcription factor A; TIF2, transcriptional intermediary factor 2; TR, thyroid hormone receptor; TZD, thiazolidinedione; UCP1, uncoupling protein 1; WAT, white adipose tissue
DISTINCTIVE FEATURES OF WHITE AND BROWN ADIPOSE TISSUES
WAT and BAT (white and brown adipose tissue respectively) are normally localized in anatomically distinct areas in rodents, although the presence of brown adipocytes in typical white fat pads occurs under certain conditions [1]. The two types of fat tissue also differ ontogenically in mice and rats, as BAT and WAT develop pre- and postnatally respectively [1]. Whereas both WAT and BAT develop before birth in other mammals, such as rabbits, cats, dogs, sheep, goats, cattle, reindeer, monkeys and humans, BAT develops postnatally in hamsters [2–4]. Several features distinguish the adipocytes of WAT and BAT. First, whereas white fat cells usually contain one major (unilocular) lipid droplet that fills up the majority of the cytoplasm, brown fat cells contain several small (multilocular) lipid droplets [1]. Secondly, brown adipocytes contain high numbers of mitochondria packed with cristae. White fat cells display relatively few mitochondria. Thirdly, BAT is highly vascularized and highly innervated by the SNS (sympathetic nervous system). The differences in lipid content and mitochondrial abundance in white and brown fat cells, as well as in vascularization of the tissues, are the reasons for the colour differences between WAT and BAT. WAT and BAT perform essentially opposite functions in vivo, with WAT accumulating excess energy as triacylglycerol and BAT dissipating energy through adaptive thermogenesis. The thermogenic capacity of BAT is due to expression of UCP1 (uncoupling protein 1) exclusively in brown adipocytes. UCP1 is a facultative proton transporter localizing to the mitochondrial inner membrane, where it can uncouple the oxidation of fuel substrates from the production of ATP, instead generating heat. In rodents, expression of UCP1 is highly responsive to external stimuli such as food intake and changes in temperature. Food intake and exposure to cold signal to BAT through SNS-derived noradrenaline (norepinephrine), which, in turn, activates ARs (adrenergic receptors) on brown fat cells. Signalling through ARs in adipocytes has been reviewed recently [2,5] and will be discussed only briefly below.
EVIDENCE FOR ANTI-OBESITY FUNCTIONS OF BAT AND UCP1
Early evidence for BAT as a tissue affecting adiposity in rodents came from experiments in which interscapular BAT was surgically denervated or excised. In some reports such animals accumulated abnormal amounts of body fat [6]. Transgenic mice expressing diphtheria toxin A from the UCP1 promoter have a 60–70% reduction in BAT mass, and these mice are obese and sensitive to both cold and diet-induced obesity when kept below thermoneutrality [7–9]. Mice lacking UCP1 are cold-sensitive, yet are neither obese nor prone to diet-induced obesity [10]. Transgenic mice expressing UCP1 from the FABP4 (fatty acid-binding protein 4; also known as aP2) promoter are resistant to genetic and diet-induced obesity [11,12]. These mice express UCP1 in WAT at levels 2–10% of that observed in BAT, but mice homozygous for the transgene are cold-sensitive due to BAT atrophy, indicating that ectopic expression of UCP1 in WAT is unable to compensate for impaired BAT-dependent thermogenesis [13]. Targeted disruption of Cidea (cell death-inducing DFF45-like effector A) results in lean mice that are resistant to diet-induced obesity [14]. Of interest, biochemical data suggest that Cidea inhibits the uncoupling activity of the UCP1 protein, possibly through a direct interaction between UCP1 and Cidea. Moreover, BAT is dysfunctional in genetically obese mouse models like ob/ob (leptin-deficient) and db/db (leptin receptor-deficient) [2,15,16]. However, whether impaired BAT function is the cause or consequence of obesity is not clear. Finally, in most mouse models described below, proneness to obesity correlates with decreased BAT activity, and resistance to obesity correlates with increased BAT function or the induction of brown adipocyte-like gene expression in WAT.
PLASTICITY OF WAT AND BAT
The developmental relationship between white and brown adipocytes is not clear. Are they derived from common or separate precursor cells? Can a white fat cell through transdifferentiation transform into a brown adipocyte and vice versa? Several studies have addressed the question of plasticity between the two types of adipocytes in vivo. As mentioned above, the anatomical location of depots of WAT and BAT is relatively distinct. In addition, it has been shown that inguinal white fat cells arise independently of the brown adipose lineage in mice kept at ambient temperature [17]. Moreover, cells in the SV (stromal-vascular) fraction (containing the preadipocytes) of WAT and BAT differentiate mainly into white and brown adipocytes respectively [18]. In rodents, BAT depots persist into adulthood, whereas BAT depots shortly after birth are replaced partially or completely by WAT in larger mammals such as cats [19], dogs [20], goats [21], sheep [22], cattle [22], reindeer [23], red deer [23] and humans [24]. In all larger mammals, UCP1 expression is most prominent in visceral depots, with particular high levels of expression observed in perirenal adipose tissue. The mechanism by which BAT transforms into WAT after birth is unknown, but might be due to transdifferentiation of brown adipocytes into white fat cells or neodifferentiation of white preadipocytes. In sheep and cattle, it was demonstrated that transcription of the UCP1 gene in visceral adipose tissues ceases abruptly at birth [22]. The BAT-to-WAT transition after birth reverts in response to cold exposure or β-adrenergic agonists in cats [25] and dogs [20,26], but not in sheep [27] and reindeer [23]. On the basis of quantification of UCP1 mRNA levels in human intraperitoneal adipose tissue, it has been estimated that 1 in 100–200 adipocytes expresses UCP1, but it is unknown to what extent these cells contribute to energy expenditure [28]. There is circumstantial evidence that cold ambient temperatures may induce the formation of adipose tissue resembling BAT around neck arteries and in the pericardium [29]. Plasma catecholamines increase during cold exposure [30], but even though some reports have indicated the existence of adaptive adrenergic non-shivering thermogenesis in humans, the contribution to overall thermogenesis is minor [2]. Significant induction of UCP1 expression in adults is associated with pathological conditions [2,4]. Thus, induction of UCP1 expression in intraabdominal adipose tissues is observed in patients with pheochromocytoma [31,32], and UCP1 is expressed in hibernomas, a BAT tumour [33]. In mice and rats, exposure to cold or β-adrenergic agonists induces the appearance of brown adipocytes in traditional white fat pads [34–37]. Interestingly, the extent varies greatly in different mouse strains, suggesting that the appearance of brown adipocytes in WAT is under genetic control. Continuous exposure for 7 days to a β-adrenergic agonist induces the appearance of multilocular, mitochondria-rich adipocytes in the retroperitoneal white fat pad of rats [37]. A subset of these multilocular adipocytes expresses UCP1. They were suggested to arise by transdifferentiation of existing white fat cells, and not by proliferation of brown adipocyte precursor cells for two reasons: first, the number of unilocular fat cells declined substantially during treatment, and secondly, the multilocular adipocytes did not incorporate bromodeoxyuridine during the course of the β-adrenergic stimulation [37]. Finally, the 3T3-L1 and 3T3-F442A mouse white preadipocyte cell lines can be induced to express UCP1 by forced expression of PGC-1α [PPARγ (peroxisome proliferator-activated receptor γ) co-activator-1α] or SV40 (simian virus 40) TAg (large T antigen) [38,39] (see below). Similarly, forced expression of PGC-1α in human white adipocytes induces expression of UCP1 [40]. Taken together, it appears that some plasticity exists between the white and brown adipocyte lineages, but the extent of such lineage shifts and the molecular mechanisms governing these remain largely unknown.
MODEL SYSTEMS TO STUDY WHITE AND BROWN ADIPOCYTE DIFFERENTIATION
Understanding the molecular regulation of adipocyte differentiation through in vivo studies is complicated by the fact that adipose precursor cells differentiate relatively asynchronously and are difficult to detect in situ. The use of SV fractions from WAT and BAT is an informative and commonly used approach. However, the low yield of cells from such isolations, the various differentiation stages displayed by the isolated cells and their limited proliferative life span in vitro necessitate frequent isolations, which makes it a laborious option. In addition, primary brown adipocytes have been reported to express very low levels of some genes characteristic of BAT, including PPARα (which in mice is expressed at the highest level in BAT of all tissues) and β2-AR [41–44]. The establishment of cell lines capable of differentiating into white or brown adipocytes has been fruitful tools in deciphering molecular aspects of adipogenesis and adipocyte function [45,46]. White preadipocyte cell lines, most prominently 3T3-L1 and 3T3-F442A (both subclones of the spontaneously immortalized Swiss 3T3 cells), have been extensively used to study white adipogenesis. Several mouse brown preadipocyte cell lines have been established, including HIB-1B (derived from a TAg-induced BAT tumour) [47], MB4 (derived from primary brown preadipocytes by forced expression of TAg and oncogenic H-Ras) [48], B7-4 (derived from a TAg-induced BAT tumour) [49] and HB2 (derived from p53−/− primary brown preadipocytes) [50]. In addition, primary brown preadipocyte cultures immortalized by TAg retain the ability to differentiate into brown adipocytes [51]. Cell lines are attractive tools due to their high proliferative capacity (as they are immortal), easy handling and reproducible differentiation; however, they too have limitations. A DNA microarray analysis demonstrated that multiple genes were differentially expressed in 3T3-L1 adipocytes compared with white adipocytes in vivo, and brown adipocytes differentiated from cell lines generally express low levels of UCP1 and PPARα compared with BAT [47,48,52–54].
DIFFERENTIAL GENE EXPRESSION IN WHITE AND BROWN PREADIPOCYTES AND ADIPOCYTES
Several established key transcriptional regulators of white adipocyte differentiation, PPARγ and members of the C/EBP (CCAAT/enhancer-binding protein) family, also regulate brown adipogenesis [55] (see below). The molecular events that govern white versus brown adipose conversion have therefore remained elusive. Nevertheless, many genes are differentially expressed in mature white and brown adipocytes. Genes involved in mitochondrial biogenesis and function are often enriched in BAT relative to WAT, whereas UCP1 is the only gene reported to display an entirely brown adipocyte-specific expression [2,56]. The functional characterization of regulatory regions of the UCP1 gene has revealed the importance of critical response elements in the proximal promoter as well as in a short distal enhancer [2]. The proximal promoter contains a binding site for CREB (cAMP response element-binding protein), and the distal enhancer contains binding sites for TRs (thyroid hormone receptors), PPARs, RXRs (retinoid X receptors), retinoic acid receptors and CREB [2]. The distal enhancer is required for BAT-specific expression of UCP1. PPARα and Dio2 (type 2 iodothyronine deiodinase) are other examples of genes highly enriched in BAT relative to WAT [57,58]. Until recently, much less was known about genes preferentially expressed in WAT, with leptin being an exception [59]. However, a DNA microarray analysis comparing gene expression in rat epididymal WAT and interscapular BAT revealed a list of such genes, with fibrillin-1, WAP four-disulfide core domain 10A and thioesterase B found to be expressed at nearly 100-fold higher levels in WAT [56]. To gain insight into the regulatory circuits controlling white versus brown adipogenesis, it is perhaps of greater interest to identify genes differentially expressed in white and brown preadipocytes, e.g. cells from SV fractions of WAT and BAT. Only few molecular markers for preadipocytes are known, and even less is known about markers that can distinguish precursor cells originating from WAT and BAT. In an attempt to address this question, differentially expressed genes of SV fractions from WAT and BAT of the Djungarian dwarf hamster were identified by representational difference analysis followed by DNA microarray analysis [60]. The result was the identification of four genes expressed at higher levels in the WAT SV fraction and seven genes expressed at higher levels in the BAT SV fraction. The four genes enriched in white preadipocytes were three complement factors (B, C2 and C3) and the Δ6 fatty acid desaturase, and the seven genes enriched in brown preadipocytes were fibronectin, α-actinin-4, metargidin, vigilin, hepatocellular carcinoma-associated protein, small nuclear ribonucleoprotein polypeptide a and necdin [60]. The differential expression of the 11 genes was not confirmed at the protein level, in SV fractions from other rodent species or in white and brown preadipocyte cell lines. In addition, except for necdin (see below), their possible involvement in white or brown adipose conversion remains to be determined. It is very likely that many more genes are differentially expressed in white and brown preadipocytes, and it would therefore be of significant interest to carefully compare suitable model systems of white and brown adipocyte differentiation, e.g. the WAT and BAT SV fractions from other species, like mouse or rat, using large-scale gene expression profiling.
MOLECULAR REGULATION OF WHITE VERSUS BROWN ADIPOCYTE DIFFERENTIATION
Despite adipogenesis being an active area of research, little is known about molecular mechanisms controlling white versus brown adipocyte differentiation. However, recent years have brought to light some examples of potential regulators, identified mainly through studies of gene-targeted mice or cells derived from gene-targeted mice. In this section, we will summarize recent advances in the understanding of factors differentially regulating white and brown fat cell differentiation, including conditions that are associated with increased energy expenditure and UCP1 expression in WAT.
βARs, Foxc2 (forkhead box C2) and cAMP sensitivity
Noradrenaline-induced activation of βARs is important for both activation of existing brown fat cells and recruitment of new brown adipocytes [2,5]. βARs are G-protein-coupled receptors that signal through an increase in intracellular cAMP and subsequent activation of PKA (cAMP-dependent protein kinase). βARs are believed mainly to couple to Gsα. Activation of existing BAT by β-adrenergic stimulation depends on the activity of the p38 MAPK (mitogen-activated protein kinase), which is activated downstream of PKA [5]. The crucial role of βARs in BAT function was confirmed by the creation of mice lacking all three βAR subtypes [41,61]. Whereas BAT of mice expressing only the β2-AR subtype had a normal morphology, brown adipocytes of mice lacking all three βARs have unilocular lipid droplets and these mice are highly sensitive to both diet-induced obesity and cold. Accordingly, they fail to induce expression of UCP1 in response to cold. In addition, increased expression of Gsα in WAT has been linked to induction of UCP1 expression, mitochondrial biogenesis and increased energy expenditure [62]. Support for a role of cAMP signalling in BAT function was suggested by targeted disruption of the PKA regulatory subunit RIIβ [63]. In brown adipocytes of these mice, a compensatory up-regulation of the RIα regulatory subunit is observed. As RIα has a higher affinity for cAMP than RIIβ, this causes the brown adipocytes to be cAMP-hypersensitive for activation of PKA. The knockout mice have diminished WAT depots, increased expression of UCP1 in BAT, and they are lean and resistant to diet-induced obesity [63]. One factor controlling the expression of RIα is the adipose tissue-enriched forkhead transcription factor Foxc2 [64]. In transgenic mice expressing Foxc2 in white and brown fat, an induction of RIα was observed and the adipocytes of WAT and BAT were hypersensitive to cAMP. In addition, a partial morphological conversion of WAT into BAT-like tissue occurred, and this BAT-like tissue expressed UCP1 and increased levels of PGC-1α. Finally, the Foxc2-transgenic mice are lean and resistant to diet-induced obesity [64]. A potential role of Foxc2 and RIα in white versus brown adipose conversion has recently been supported by in vitro experiments [39]. However, the physiological role of Foxc2 in WAT and BAT is not fully understood for several reasons. First, Foxc2 was shown to be expressed at higher levels in the adipose compared with the SV fraction, but paradoxically to be down-regulated during the differentiation of 3T3-L1 preadipocytes and MEFs (mouse embryo fibroblasts) [39,64,65]. Secondly, Foxc2 was reported to be expressed at equal levels in WAT and BAT [64], whereas others have found substantially lower levels of Foxc2 in the latter [66]. Thirdly, recent data demonstrate that forced expression of Foxc2 blocks adipose conversion of 3T3-L1 preadipocytes and overrides the ability of retrovirally expressed PPARγ to induce differentiation of non-adipose fibroblasts [67].
The PGC-1 family
PGC-1α was cloned as a PPARγ-interacting protein with an enriched expression in BAT compared with WAT [38]. It interacts with numerous nuclear receptors and is strongly induced in BAT in response to cold exposure. In addition, PGC-1α potently co-activates both PPARγ and TRβ on the UCP1 promoter [38]. Forced expression of PGC-1α in white fat cells induces mitochondrial biogenesis and expression of UCP1 [38,40,68]. A mechanism through which PGC-1α induces mitochondrial biogenesis was subsequently demonstrated in myoblasts [68]. In these cells, PGC-1α induces expression of NRF-1 (nuclear respiratory factor 1) and NRF-2α/GABPα (GA-binding protein α) and functions as a co-activator of NRF-1 on the Tfam (mitochondrial transcription factor A) promoter [68]. NRFs are important transcriptional regulators of many respiratory chain genes and Tfam is a crucial regulator in transcription and replication of the mitochondrial genome [69]. The mechanism through which PGC-1α regulates mitochondrial biogenesis in brown adipocytes has not been resolved, although it is expected to be similar to that in myoblasts. Another potentially important factor mediating the effects of PGC-1α on mitochondrial biogenesis and respiration is ERRα (oestrogen-related receptor α) [70,71]. The link to ERRα has, however, not been established in brown adipocytes. Expression of PGC-1α is strongly induced during brown adipocyte differentiation [39,72], yet little is known about the regulators of PGC-1α expression in this setting. In hepatocytes and myoblasts, the PGC-1α promoter is positively regulated by CREB (a PKA substrate) [73,74], suggesting that cAMP signalling may be involved in the induction of PGC-1α expression also during brown adipogenesis. Together, these data point to a key function of PGC-1α in orchestrating UCP1 expression and mitochondrial respiration and biogenesis during brown adipose conversion as well as in mature brown fat cells. Two independent lines of PGC-1α knockout mice were reported recently [75,76]. One of these lines have impaired BAT function, with brown adipocytes displaying increased accumulation of large lipid droplets [75]. In addition, these PGC-1α knockout mice are cold-sensitive, a characteristic possibly linked to a blunted induction of UCP1 expression and other genes involved in the thermogenic response [75]. It is noteworthy that, despite impaired BAT function, these mice are resistant to diet-induced obesity due to their physical hyperactivity [75]. In the second line of PGC-1α knockout mice, however, no BAT phenotype was observed [76]. Although young PGC-1α-deficient mice were found to be cold-sensitive, this was not associated with impaired induction of UCP1 expression. Moreover, Leone et al. [76] reported a male-specific resistance to diet-induced obesity, yet this was apparently not linked to hyperactivity. The reason for the differences in BAT phenotype of the two PGC-1α knockout lines is not clear, but may relate to differences in strain background. The function of PGC-1α is far from restricted to brown fat cells, as PGC-1α has important roles in, for example, the regulation of hepatic gluconeogenesis and muscle fibre type switching [77].
Phosphorylation of three amino acid residues in PGC-1α by the p38 MAPK results in an increased stability and transcriptional activity [78]. The functionality of these phosphorylations has been addressed in brown adipocytes, in which β-adrenergic stimulation induces p38 MAPK activity [79,80]. Inhibition of p38 MAPK activation abrogates cold-induced induction of UCP1 expression in BAT as well as β-adrenergic stimulation of UCP1 expression in brown fat cells in vitro. The β-adrenergic stimulation of UCP1 expression in primary brown adipocytes is blunted in cells expressing a non-phosphorylatable PGC-1α mutant compared with cells expressing wild-type PGC-1α [79]. The mechanism by which phosphorylation activates the transcriptional activity of PGC-1α possibly relates to alleviation of the function of the repressive domain, a negative regulatory domain identified in PGC-1α [78,81]. The repressive domain in PGC-1α interacts with several proteins and these interactions are abrogated upon p38 MAPK-mediated phosphorylation [82–84]. One of the identified interacting proteins, p160 myb binding protein, has intrinsic transcriptional repressor activity and antagonizes PGC-1α-induced mitochondrial respiration [82]. Recently, two additional types of post-translational modifications were shown to influence PGC-1α activity. The activity of PGC-1α was reported to be enhanced by methylation of three arginine residues in its C-terminal region [85]. The modifications were carried out by PRMT1 (protein arginine methyltransferase 1), and they were shown to be important for activation of PGC-1α target genes involved in mitochondrial biogenesis. Moreover, PGC-1α was shown to be acetylated at multiple lysine residues, possibly in part by p300 [86,87]. The sirtuin family member SIRT1 (see below) deacetylates PGC-1α [86,87], which affects the regulation of gluconeogenic genes by PGC-1α in liver [87]. PGC-1α can also be activated by docking of transcription factors on its N-terminal domain, resulting in an altered conformation and increased interaction with other co-activators, including SRC-1 (steroid receptor co-activator 1) [81].
Two additional PGC-1α-related co-activators have been cloned [88–90]. PGC-1β shares sequence homology with PGC-1α throughout the molecule, whereas the sequence of PRC (PGC-1-related co-activator) is similar to PGC-1α mainly in the N- and C-terminal regions. The PGC-1 family members share some similarities in expression patterns and function, but differ in other aspects. Like PGC-1α, PGC-1β is highly enriched in BAT compared with WAT and powerfully induces mitochondrial respiration and biogenesis in muscle cells [88,91,92]. The expression of PGC-1β is induced during white as well as during brown adipocyte differentiation [39,72,88,93], although the level of induction during brown adipogenesis is substantially higher [39]. Of interest, PGC-1β appears to be less ‘promiscuous’ in the selection of transcription factors that it co-activates than PGC-1α [89,93]. However, all three PGC-1 family members are potent co-activators of NRF-1 [68,88,90]. Using immortalized brown preadipocytes from wild-type or PGC-1α knockout mice in which expression of PGC-1β could be silenced by RNA interference, it was shown that neither PGC-1α nor PGC-1β was necessary for brown adipocyte differentiation in vitro, including expression of UCP1 and Cidea [72]. Moreover, brown adipocytes lacking PGC-1α or PGC-1β displayed only subtle reductions in oxygen consumption and differentiation-dependent mitochondrial biogenesis. However, PGC-1α-deficient adipocytes failed to properly induce thermogenesis and thermogenic gene expression in response to cAMP. Silencing of PGC-1β expression in PGC-1α−/− preadipocytes did not interfere with differentiation per se, as evidenced by normal FABP4 expression and lipid accumulation, but severely attenuated expression of brown adipocyte-selective genes like UCP1, oxygen consumption and differentiation-dependent mitochondrial biogenesis [72]. These studies demonstrate that PGC-1α and PGC-1β are functionally redundant in brown adipose conversion, but that their combined loss compromises brown adipogenesis in vitro. PRC is ubiquitously expressed and its expression is not enriched in BAT relative to WAT [90]. In addition, neither PRC nor PGC-1β are induced in BAT after cold exposure [88,90]. It has not yet been examined whether PRC can induce mitochondrial biogenesis, although it has been linked to mitochondrial respiration and biogenesis in thyroid oncocytoma and serum-stimulated BALB/3T3 fibroblasts [94,95].
The p160 family
The p160 family of nuclear receptor co-activators comprises SRC-1, TIF2 (transcriptional intermediary factor 2) and p/CIP [p300/CBP (CREB-binding protein)/co-integrator-associated protein]. A comparison of TIF2−/− and SRC-1−/− mice revealed that TIF2−/− mice were protected against diet-induced obesity and exhibited enhanced adaptive thermogenesis, whereas SRC-1-deficient mice displayed an impaired ability for energy expenditure and adaptive thermogenesis, and furthermore, they were prone to obesity [96]. TIF2−/− MEFs had an impaired adipogenic potential, and in 3T3-L1 preadipocytes forced expression of TIF2, but not SRC-1, enhanced adipogenesis. The interscapular BAT of TIF2−/− mice had enhanced expression of UCP1, PGC-1α and acetyl-CoA oxidase. Furthermore, enlarged mitochondria with more cristae were observed, and BAT in TIF2-deficient mice contained smaller lipid droplets. As discussed above, PGC-1α is a key regulator of BAT development and UCP1 expression and stimulates transcription in part through PPARγ-dependent recruitment of co-activators harbouring histone acetyltransferase activity, including SRC-1 [81,97]. In keeping with the increased expression of UCP1 in BAT of TIF2−/− mice, SRC-1 stimulated PPARγ- and PGC-1α-dependent transactivation, and TIF2 dose-dependently attenuated the effect of SRC-1. It was noteworthy that high-fat feeding increased the level of TIF2 selectively in WAT and BAT, thus promoting fat accumulation in these two tissues. Hence, the balance between SRC-1 and TIF2 seems to play an important role in energy homoeostasis via differential regulation of the physiology of WAT and BAT, where a high ratio between SRC-1 and TIF2 favours high energy expenditure in adipose tissue. Given the observation that β-adrenergic induction of cAMP production is a key regulator of UCP1 expression in BAT and that decreased levels of TIF2 tend to enhance the activity of the PPARγ–PGC-1α complex, it is noteworthy that activation of PKA directs TIF2 to proteasome-mediated degradation [98].
By crossing SRC-1−/− and p/CIP−/− mice it was recently shown that mice lacking both co-activators exhibited a defective development of BAT [99]. The amount of BAT in double knockout mice was significantly reduced compared with both wild-type mice and single knockout mice. In double knockout mice the development of BAT was arrested at an early stage prior to the accumulation of fat. Using preadipocyte cell lines established from the immature BAT depot of the double knockout mice it was shown that the defect in BAT formation was cell-autonomous and occurred at a stage after PPARγ induction. Combined expression of p/CIP and SRC-1 rescued differentiation, and similarly, expression of a constitutively active PPARγ–VP16 fusion protein, but not wild-type PPARγ, rescued differentiation of the SRC-1−/−/p/CIP−/− preadipocytes. The effect of lack of SRC-1 and p/CIP on gene expression was selective, as expression of several genes characteristic of adipose tissue was unaffected, whereas particularly expression of UCP1 was down-regulated. ChIP (chromatin immunoprecipitation) analysis of BAT from newborn double knockout mice demonstrated decreased binding of PPARγ to the UCP1 enhancer region. Furthermore, binding of CBP and TIF2 was also decreased, whereas binding of the repressors RIP140 (receptor interacting protein 140) and NCoR (nuclear receptor co-repressor) to the UCP1 enhancer region was increased. Thus, in the absence of SRC-1 and p/CIP, impaired recruitment of PPARγ to the UCP1 enhancer combined with decreased recruitment of certain co-activators and enhanced recruitment of co-repressors block proper development of functional BAT, and accordingly the double knockout mice exhibit defective adaptive thermogenic responses to cold and high-fat feeding. Yet it should be noted that the double knockout mice remain lean as a result of an increased basal metabolic rate and higher physical activity [99].
RIP140
RIP140 is an unusual co-repressor that is recruited to most nuclear receptors in a ligand-dependent manner [100–102]. RIP140 is expressed in most tissues, but within one tissue, expression is often confined to specific cell types. High expression of RIP140 is observed in gonadal tissues, WAT and muscle, whereas expression in BAT is lower [66,103]. RIP140 knockout mice are viable, but females are infertile as a result of impaired ovulation [104]. Interestingly, RIP140 knockout mice display a lean phenotype with a decrease in total body fat of about 70%. RIP140−/− mice are almost completely devoid of subcutaneous WAT and exhibit a marked decrease in other WAT depots. Histological examinations revealed that the reduction of adipose mass was due to a decreased content of triacylglycerol and not adipocyte cell number. Food intake was not diminished and physical activity was not increased. The decrease in adipose mass was not accompanied by accumulation of triacylglycerol in other tissues, and insulin-sensitivity appeared normal. Finally, RIP140 mice gained less weight on a high-fat diet than normal mice [66]. These findings suggested that energy expenditure was increased in RIP140 knockout mice, and accordingly, expression of genes involved in mitochondrial β-oxidation was up-regulated in RIP140 knock-out mice, and, even more conspicuously, a greater than 100-fold up-regulation of UCP1 expression was observed in WAT. Surprisingly, no increase in PGC-1α mRNA expression was observed, and likewise, the levels of SRC-1 and TIF2 were unaltered [66]. To distinguish systemic effects from cell autonomous effects, the role of RIP140 was investigated in primary cultures of WAT and in MEFs. These analyses demonstrated that expression of RIP140 increases during the process of adipocyte differentiation, but RIP140 is not essential for adipogenesis per se [66,105]. In the absence of RIP140, expression of genes involved in energy expenditure, including UCP1, Cidea and CPT-1b (carnitine palmitoyltransferase 1b) was increased, and forced expression of RIP140 in RIP140-deficient cells prevented this increase. In keeping with the increased expression of UCP1 and genes facilitating fatty acid oxidation, the rate of β-oxidation was increased in the RIP140−/− cells [105]. Thus, RIP140 cell autonomously controls expression of key genes involved in energy expenditure, and using ChIP, it was shown that RIP140 is targeted to the UCP1 enhancer, which, as mentioned, harbours binding sites for PPARs and TRs [105]. A recent study furthermore demonstrated that RIP140 in an ERRα-dependent manner functions as a global regulator of genes involved in glucose uptake, glycolysis, the tricarboxylic acid cycle, fatty acid oxidation and oxidative phosphorylation, and that ablation of RIP140 increased insulin sensitivity and glucose tolerance [106]. As mentioned, expression of RIP140 is lower in BAT than in WAT and it is conceivable that this at least in part determines UCP1 expression. As mentioned, despite increased UCP1 levels, the expression of PGC-1α and PGC-1β mRNAs was not increased in RIP140-deficient adipose tissue and cell lines. Consistently, mitochondrial numbers were not altered in RIP140-deficient adipocytes.
Thyroid hormone, TRs and bile acids
The function of thyroid hormone in energy homoeostasis has been intensely studied and is very important, yet it is still incompletely understood [2]. Thyroid hormone action is mediated by the family of TRs, which can function as transcriptional repressors or activators in the absence or presence of thyroid hormone respectively. Studies addressing the function of thyroid hormone in adipose tissue have focused on its role in BAT. The UCP1 gene is thyroid hormone-responsive, and thyroid hormone is often used for differentiation of brown preadipocytes in vitro [47,107]. In addition, activation of BAT by cold exposure causes a dramatic increase of the expression and activity of Dio2, which catalyses the conversion of T4 (thyroxine) into the active substance T3 (3,5,3′-tri-iodothyronine) [108]. In this way, brown adipocytes boost the local concentration of T3, thereby saturating their TRs, and they serve as a source of systemic T3 [2,108]. In mice, bile acids induce expression of Dio2, PGC-1α and UCP1 in BAT, and they prevent and reverse high fat-diet induced weight gain [109]. The effects of bile acids on body weight are dependent on Dio2, as they are not observed in Dio2-deficient mice. Mechanistically, bile acids induce Dio2 expression by increasing cAMP production in brown fat cells through the G-protein-coupled receptor TGR5 (G-protein-coupled bile acid receptor 1) [109]. Using different mouse strains with dramatically varying abilities to induce brown adipocyte differentiation and UCP1 expression in retroperitoneal WAT in response to cold exposure, it was found that expression of Dio2 was linked to UCP1 expression [110]. This observation indicates a possible role for Dio2, and thereby T3, in controlling the appearance of brown adipocytes in WAT. Surprisingly, mice lacking all T3-binding TRs (i.e. TRα1 and TRβ) have normal levels of UCP1 expression under thermoneutral conditions, and apparently normal BAT recruitment and induction of UCP1 expression in response to cold [111]. However, for unknown reasons, these mice are cold-sensitive. Therefore, although thyroid hormone influences BAT function, the mechanism through which this happens is not yet fully explored.
The PPAR family and PPARγ ligands
PPARγ is the master regulator of adipose conversion in vitro, being necessary as well as sufficient for adipogenesis [55,112,113]. Accordingly, PPARγ is required for the formation of both WAT and BAT in vivo, as demonstrated by exclusion of PPARγ−/− ES (embryonic stem) cell-derived cells from the adipose fraction of chimaeric mice [114], tetraploid rescue of a PPARγ-deficient embryo [115], fat-specific deletion of PPARγ [116,117], generation of mice homozygous for a hypomorphic PPARγ allele [118] and by deletion of the PPARγ2-specific exon B [119]. In addition, inducible ablation of PPARγ in mature white and brown adipocytes has shown that PPARγ is required for their survival [120].
PPARγ ligands have significant impact on adipocytes as well as adipose tissue. TZDs (thiazolidinediones) are ligands for PPARγ, and they powerfully promote white and brown adipogenesis in vitro [121,122]. Moreover, they increase expression of UCP1 in cultured brown adipocytes [122–124]. TZD as well as non-TZD PPARγ ligands can induce UCP1 expression in BAT [124–126], BAT hyperplasia [122,125] as well as the appearance of brown adipocytes and UCP1 expression in WAT depots [126–128]. Strikingly, TZD treatment induces mitochondrial biogenesis in white adipocytes in vitro and in vivo and results in mitochondrial remodelling to a cristae-rich morphology [128,129]. This effect is possibly due to the ability of TZDs to induce expression of PGC-1α in white and brown adipocytes in vitro and in vivo [126,128,130,131]. It is generally acknowledged that selective recruitment of coactivators either caused by tissue- or cell-specific differences in expression levels or as a result of the binding of selective receptor modulators dictates the transcriptional response [132]. A number of SPPARMs (selective PPARγ modulators) have been developed recently [133–136]. Common to these SPPARMs is their reduced adipogenic activity in comparison with classical TZDs. One of these, Fmoc (fluoren-9-ylmethoxycarbonyl)-L-leucine, was shown to preferentially recruit SRC-1, but not TIF2 in glutathione S-transferase pull-down experiments and enhance PPARγ-dependent transactivation in the presence of SRC-1, but not TIF2 or p300 [133]. This is a potentially important observation due to the phenotypes of mice lacking SRC-1 or TIF2 (see above).
As expression of PPARα is very high in BAT and low in WAT, a function of PPARα in brown adipocyte differentiation and function has been expected. In reporter assays, PPARα is clearly capable of activating the UCP1 promoter in a manner potentiated by PGC-1α-mediated coactivation [137–139]. In addition, a genetic analysis of different mouse strains linked PPARα to appearance of brown adipocytes and UCP1 expression in retroperitoneal WAT [110]. Finally, PPARα ligands have in some studies been reported to induce UCP1 expression in primary brown adipocytes and in BAT [110,138,140]. However, others have failed to detect induction of UCP1 expression in response to PPARα ligand in either primary brown adipocytes or BAT [125,137,141]. Moreover, analyses of PPARα−/− mice have demonstrated that the morphology of BAT appears normal, that expression of UCP1 is normal and that they are not cold-sensitive [110,140,142,143]. Therefore, the exact function and importance of PPARα in brown adipocytes are presently unclear.
Whereas PPARδ clearly plays an important role in skeletal muscle, where activation or forced expression controls oxidative metabolism and development [144–146], the role of PPARδ in adipose biology has been a matter of dispute. Initial experiments indicated that PPARδ was not adipogenic [147]. Now it appears that PPARδ controls preadipocyte proliferation [148,149] and its activation may induce PPARγ expression [149–152]. Forced expression of a constitutively active form of PPARδ in adipose tissue resulted in animals with markedly less WAT, which was attributed to less accumulation of triacylglycerol rather than a decreased number of cells [153]. BAT also contained less triacylglycerol, but the size of the interscapular BAT was not reduced. In the transgenic mice, an increased expression of genes involved in fatty acid oxidation and energy dissipation was observed in BAT and WAT. Specifically, it was shown that UCP1 was expressed in WAT of the transgenic mice, even though the level of expression was significantly lower than in BAT. In keeping with the altered gene expression profile, the transgenic mice were protected against high-fat diet-induced obesity. As expression of constitutively active PPARδ also counteracted weight gain in db/db mice, it was concluded that the effect did not require leptin signalling. Furthermore, treatment of db/db mice with the potent PPARδ agonist GW501516 for 7 days decreased lipid accumulation in BAT. Using co-immunoprecipitation, PPARδ was shown to interact with PGC-1α in a manner that was enhanced by GW501516 and dependent on the LXXLL motif located between amino acids 142 and 146 of PGC-1α. Furthermore, it was shown that PGC-1α was a powerful activator of ligand-dependent as well as ligand-independent PPARδ-mediated transactivation, and it was suggested that PPARδ via PGC-1α could regulate UCP1 expression and, hence, increase thermogenesis. Finally, it was demonstrated that high-fat feeding-induced UCP1 expression was severely compromised in adipose-specific PPARδ knockout mice [153]. This last observation clearly suggest that PPARδ may play a role in diet-induced adaptive thermogenesis, but as the phenotype of the adipose-specific PPARδ knockout is rather complex [154], the biological significance of PPARδ in relation to thermogenesis remains to be fully established.
LXR (liver X receptor)
LXRs are established regulators of cholesterol, lipid and glucose homoeostasis [155]. LXRα and LXRβ are highly expressed in adipose tissue, LXRβ being constitutively expressed in preadipocytes and adipocytes, whereas expression of LXRα is strongly induced during adipocyte differentiation [156,157]. However, the role of LXR in adipocyte differentiation and function has been ambiguous. No adipose phenotype was reported for LXRα-deficient mice [158], whereas it was reported that adipose depots were smaller in LXRα−/−/LXRβ−/− double knockout mice [157]. Administration of synthetic LXR agonists has been reported to have no effect on adipogenesis [156,159] or stimulate adipogenesis in vitro [160]. In mature adipocytes, activation of LXR induces expression of genes involved in lipid and glucose homoeostasis [156,159,161–164], and it was reported that activation of LXR enhanced triacylglycerol accumulation in adipocytes [157,160]. In keeping with an effect of LXR activation on lipid accumulation in adipocytes, it was recently shown that LXRβ is required for the age-dependent or diet-induced increase in adipocyte size [165]. In the same study it was found that UCP1 expression was markedly increased in gonadal adipose tissue of LXRβ-deficient mice. However, no marked differences in daily food intake or oxygen consumption between wild-type and LXRβ−/− mice were observed [165]. Unexpectedly, LXRα−/−/LXRβ−/− mice were found to be resistant to diet-induced obesity when fed a western high-fat high-cholesterol diet, but not when fed a cholesterol-free high-fat diet [166]. The double knockout mice have a defective hepatic lipid metabolism, but impaired SREBP-1c (sterol regulatory element-binding protein 1c)-dependent lipogenesis did not explain the observed obesity resistance. Rather the LXRα−/−/LXRβ−/− mice exhibited enhanced energy dissipation due to ectopic expression of UCP1 in WAT and muscle. Expression of Dio2 was strongly up-regulated in liver, but no increase in plasma T3 levels was observed, making it unlikely that T3 affected UCP1 expression in muscle and adipose tissue. Interestingly, induction of UCP1 expression was not paralleled by enhanced expression of PGC-1α mRNA. PGC-1α protein levels were not determined. The notion that LXRs, particularly LXRβ, may play a role in controlling UCP1 expression is consistent with the finding that administration of LXR agonist to mice suppresses UCP1 expression in BAT [167].
SHP (small heterodimer partner)
SHP is an unusual orphan nuclear receptor in that it lacks a DNA-binding domain [168]. It normally functions as a repressor and interacts with numerous other nuclear receptors, including LRH-1 (liver receptor homologue 1), LXRs, ERRs, PPARs and RXRs. The best characterized function of SHP is its involvement in feedback regulation of bile acid production in liver through repression of LRH-1 activity [168]. Recently, it was reported that SHP inhibits expression of PGC-1α in brown adipocytes [169]. Mice lacking SHP are resistant to diet-induced obesity, mainly due to diminished accumulation of triacylglycerol in adipose tissue. SHP-deficient mice display increased energy expenditure linked to altered thermogenic gene expression, including increased expression of PGC-1α and UCP1 in WAT and BAT [169]. Using primary cells, increased expression of PGC-1α and UCP1 was observed in SHP−/− brown adipocytes. Accordingly, silencing SHP expression in wild-type brown adipocytes resulted in higher levels of PGC-1α expression, whereas reintroduction of SHP cDNA into SHP−/− brown fat cells led to decreased expression of PGC-1α and UCP1 [169]. SHP was found to interact with a region of the PGC-1α promoter containing a putative ERR binding site as well as to repress ERRγ-dependent activation of the PGC-1α promoter in a dose-dependent manner. Although the expression pattern of SHP during white and brown adipocyte differentiation is not known, expression of SHP was down-regulated in BAT upon cold exposure or acute β-adrenergic stimulation [169], suggesting a physiological function of SHP in the regulation of energy expenditure in BAT.
The C/EBP family
Three members of the C/EBP family, C/EBPβ, C/EBPδ and C/EBPα, play important roles in adipocyte differentiation [55,170]. The early regulators C/EBPβ and C/EBPβ are involved in the formation of both WAT and BAT as well as in white adipocyte differentiation in vitro [171,172]. Mice lacking C/EBPβ display reduced depots of both WAT and BAT, a phenotype exacerbated in C/EBPβ−/−/C/EBPδ−/− mice. Moreover, UCP1 expression and lipid accumulation are decreased in embryonic day 18.5 BAT of knockout embryos, particularly in double knockouts [172]. Interestingly, expression of adipocyte marker genes in the WAT present in adult mice lacking C/EBPβ and/or C/EBPδ is unaltered compared with wild-type mice [172]. Similar results were obtained upon analysis of adult BAT from C/EBPβ-deficient mice [171]. Likewise, induction of PGC-1α and UCP1 expression was normal in C/EBPβ−/− mice in response to cold, although they were cold-sensitive, possibly due to impaired lipid metabolism. Finally, primary brown preadipocytes lacking C/EBPβ differentiated normally in vitro, and the primary adipocytes expressed elevated basal levels of UCP1 as well as elevated levels of PGC-1α and UCP1 after noradrenaline stimulation [171]. Thus C/EBPβ and C/EBPδ appear to play an important role during the development of WAT and BAT, but to play minor roles in mature adipose tissue.
C/EBPα knockout mice die from hypoglycaemia within 12 h after birth, but display absence of lipid accumulation in BAT [173,174]. Moreover, detailed analysis of pre- and neonatal BAT demonstrated that C/EBPα-deficiency results in decreased and/or delayed expression of PPARγ, UCP1, PGC-1α, PPARα, NRF-2 and TRs, attenuated Dio2 activity as well as defective mitochondrial biogenesis and maturation [175]. C/EBPα−/− mice transgenic for C/EBPα in liver have an improved neonatal survival compared with non-transgenic C/EBPα knockout animals [176]. At 7 days of age, BAT of transgenic C/EBPα null mice is largely indistinguishable from that of corresponding wild-type mice with respect to mitochondrial numbers and morphology, as well as expression of UCP1 and several adipocyte markers [175,176]. A few differences were noticed, though, including a tendency of C/EBPα-deficient brown adipocytes to accumulate larger lipid droplets and to express higher levels of C/EBPδ, possibly as a compensatory mechanism. Although the C/EBPα transgene rescued the BAT phenotype, this was not the case for WAT [176]. All WAT depots were still absent, except for the mammary fat pad. These data suggest that the brown fat phenotype described in C/EBPα-deficient mice is non-cell autonomous and secondary to other functions of C/EBPα, possibly as a regulator of gluconeogenesis, whereas the defective development of white adipocytes is cell autonomous. Consistently, postnatal ablation of C/EBPα selectively affects WAT and not BAT [177]. An important function of C/EBPα in promoting white adipogenesis in vitro and in vivo appears to be repression of E2F-dependent transcription [178]. Mice homozygous for a knockin of C/EBPβ in the C/EBPα locus display elevated amounts of interscapular BAT and severely decreased amounts of WAT in which the fat cells morphologically resemble brown adipocytes [62,179]. Moreover, adipocytes from homozygous knockin WAT express UCP1, contain higher levels of cAMP and possess more mitochondria, which in addition to being more numerous also are cristae-rich [62]. In summary, it appears that C/EBPα is strictly required for white adipogenesis in vivo, whereas it is dispensable for brown adipose conversion.
Insulin receptor substrates and necdin
Insulin and IGF-1 (insulin-like growth factor 1) signal through activation of their respective cell surface receptors, which trigger phosphorylation of intracellular IRSs (insulin receptor substrates) mediating a plethora of downstream signalling events [180]. Insulin/IGF-1 signalling strongly promotes adipocyte differentiation [45]. Consistently, mice lacking IRS-1 have reduced amounts of WAT, a phenotype exacerbated in mice lacking both IRS-1 and IRS-3 [181]. Moreover, a subset of IRS proteins is required for adipose conversion of SV40 TAg-immortalized brown preadipocyte cell lines, as demonstrated by the use of cells prepared from mice carrying targeted disruptions of individual or combinations of IRS protein-encoding genes [182]. Using this model system, it was shown that IRS-1-deficient cells were strongly impaired in brown adipogenesis, whereas cells lacking IRS-3 were marginally impaired. The combined absence of IRS-1 and IRS-3 fully blocked differentiation. Cells lacking IRS-2 or IRS-4 differentiated as efficiently as wild-type cells [182]. Comparison of microarray expression data from the same preadipocytes lacking individual IRS proteins demonstrated that expression of a panel of genes, including necdin, was negatively correlated with the ability to differentiate into brown adipocytes [183]. Necdin was expressed at 40-fold higher levels in IRS-1-deficient brown preadipocytes compared with wild-type preadipocytes, and this was partially reversed upon reintroduction of IRS-1 cDNA into IRS-1-deficient cells. Intriguingly, silencing of necdin expression by RNA interference in cells lacking IRS-1 partially restored differentiation and expression of marker genes like PPARγ, PGC-1α and UCP1 [183]. Thus necdin appears to be an inhibitor of brown adipogenesis, at least of immortalized IRS-1−/− preadipocytes. However, ectopic expression of necdin in wild-type brown preadipocytes to levels observed in cells lacking IRS-1 has no impact on either differentiation or BAT-specific gene expression [183]. In addition, whereas WAT is strongly affected in mice lacking IRS-1, BAT apparently is unaffected, even in mice lacking both IRS-1 and IRS-3 [181]. Moreover, the differential expression of necdin in immortalized wild-type and IRS-1-deficient brown preadipocytes is not recapitulated in vivo, as necdin is not expressed at significantly different levels in BAT from wild-type and IRS-1-deficient mice [183]. Finally, how the observation that necdin is expressed at higher levels in SV fractions from BAT than in SV fractions from WAT [60] relates to the observations in wild-type and IRS-1−/− brown preadipocytes is unclear at present. Clearly, IRS-1, IRS-3 and necdin are potential regulators of brown adipocyte differentiation in vitro. It will be interesting to decipher their impact on brown adipose conversion in vivo, as the characterization of mice lacking IRS-1 and/or IRS-3 suggests that the IRS proteins are more important for the formation of WAT than BAT. Therefore, more studies are required to establish if IRS proteins and necdin are molecular determinants of white versus brown adipocyte differentiation.
S6K1 [40S ribosomal protein S6 kinase (p70S6K/p85S6K)]
S6K1 is a key regulator of cellular proliferation [184,185]. S6K1 is activated by mTOR (mammalian target of rapamycin), which serves as an integrator of proliferative, hormonal and nutritional signals [186]. Recently, a homologous rapamycin-sensitive S6 kinase termed S6K2 was identified [187]. Disruption of either the S6K1 gene or the S6K2 gene does not affect viability or fertility [184,187]. However, S6K1-deficient mice were shown to be hypoinsulinaemic and glucose intolerant due to a selective decrease in pancreatic β-cell mass, but surprisingly, the S6K1-deficient mice maintained normal fasting plasma glucose levels, suggesting hypersensitivity to insulin [188]. In a subsequent study S6K1−/− mice were shown to be protected against diet-induced or genetic obesity [189]. In S6K1−/− mice a marked decrease in the size of adipose depots was observed, and epididymal WAT acquired a multilocular, BAT-like appearance. Energy expenditure as determined by oxygen consumption was increased, but the fact that circulating triacylglycerol and fatty acids were similar in knockout and wild-type animals indicated that triacylglycerol and fatty acids were oxidized in adipose tissue or muscle. Accordingly, PGC-1α and UCP1 were expressed in WAT, and expression of PPARδ and PGC-1α was increased in muscle [189]. Acquisition of a BAT-like phenotype with expression of UCP1 in WAT of S6K1−/− mice probably reflects enhanced insulin/IGF-1 signalling in the absence of the negative mTOR/S6K1 feedback loop impinging on IRS-1 and IRS-2 [190]. Insulin and, in particular IGF-1, have been demonstrated to play crucial roles in adipogenic and thermogenic differentiation of foetal brown adipocytes [191–194]. Induction of UCP1 expression in differentiating brown adipocytes is dependent on an IGF-1R (IGF-1 receptor)-dependent IRS-1/IRS-2-mediated activation of Akt/PKB (protein kinase B) [194,195]. IGF-1-dependent activation of the UCP1 promoter is not inhibited by rapamycin in transient transfection experiments, and rapamycin only partly inhibited IGF-1-induced UCP1 expression in foetal brown adipocytes [196]. Furthermore, insulin-mediated S6K activation is not impaired in IGF-1R-deficient brown adipocytes, in which insulin does not induce UCP1 expression [194]. Thus, even though adipogenesis per se is inhibited by rapamycin [197], it appears that enhanced insulin/IGF-1 signalling in the absence of negative S6K feedback regulation promotes differentiation of BAT-like adipocytes with UCP1 expression.
4E-BP1 (4E-binding protein 1)
Targeted disruption of the eukaryotic translation initiation factor 4E-BP1 resulted in mice with a more than 50% decrease in WAT mass [198]. Moreover, multilocular adipocytes appeared in the inguinal and retroperitoneal WAT of knockout mice. In addition, UCP1 expression was substantially increased in inguinal WAT, confirming an increased presence of brown adipocytes, and knockout mice had an increased metabolic rate [198]. Intriguingly, whereas no difference in expression of PGC-1α mRNA in inguinal WAT was observed between wild-type and knockout mice, the level of PGC-1α protein was 2–3-fold higher in the knockout adipose tissue. Cold exposure or injection of a β-adrenergic agonist led to a rapid down-regulation of 4E-BP1 expression in BAT of wild-type mice, concomitant with an induction of UCP1 expression [199]. The cold-induced down-regulation of 4E-BP1 was absent in mice lacking β-adrenergic receptors [199]. In these studies, the level of PGC-1α was not addressed. Although speculative, it appears that 4E-BP1 can function as an inhibitor of PGC-1α mRNA translation and is thereby a potential inhibitor of brown adipogenesis.
The pRB (retinoblastoma protein) family
pRB is a tumour suppressor and a key regulator of the cell cycle and differentiation of many cell types [200]. Moreover, it is the founding member of a family also encompassing p107 and p130. The involvement of the pRB family in adipocyte differentiation has recently been reviewed [201,202]. The ability of TAg to inhibit white adipocyte differentiation is well established [203,204]. However, as mentioned above, most brown preadipocyte cell lines have been established either from TAg-expressing BAT tumours or by forced expression of TAg in primary brown preadipocytes. Moreover, transgenic mice expressing TAg from the FABP4 promoter exhibit a partial conversion of white into brown adipose tissue [47]. These results indicate a differential effect of TAg on white and brown adipogenesis. Important targets of TAg are the three pRB family members that are functionally inactivated by TAg through direct interaction [205]. Expression of TAg in several white preadipocyte cell lines resulted in expression of UCP1 in the adipose state [39]. It is noteworthy that adipose conversion of these TAg-expressing cells was obtained by addition of exogenous PPARγ ligand, which bypassed the differentiation-inhibitory effect of TAg normally observed in these cells. A point mutant of TAg that failed to bind pRB family members did not result in the induction of UCP1 expression. Mouse ES cells lacking pRB directed to differentiate into adipocytes expressed UCP1, whereas this was not the case for wild-type cells or cells lacking both p107 and p130 [39]. Consistently, pRB-deficient and wild-type MEFs differentiated into brown and white fat cells respectively. pRB-deficient adipocytes expressed UCP1 protein, as well as PGC-1α and PGC-1β mRNAs, at levels comparable with those in BAT and they contained many more mitochondria than wild-type fat cells [39]. Moreover, wild-type and pRB-deficient fat cells expressed the three βARs at levels similar to those in WAT and BAT respectively. Additional evidence that pRB regulates white versus brown adipocyte differentiation was obtained by studying primary adult white preadipocytes from mice carrying two floxed Rb (retinoblastoma gene) alleles. Cells made pRB-deficient through infection with adenovirus expressing Cre recombinase differentiated into brown fat cells, whereas cells infected with control adenovirus differentiated into white adipocytes [206]. These data establish pRB as a molecular switch involved in the commitment of precursor cells to become either white or brown adipocytes in vitro. Besides the phenotype of FABP4-TAg transgenic mice, three observations indicate a role of pRB in determining white versus brown adipose conversion in vivo: pRB is not present in preadipocytes of interscapular BAT, but is present in preadipocytes of epididymal WAT (it is noteworthy that pRB is abundantly expressed in both mature BAT and WAT); pRB levels decrease during transdifferentiation of retroperitoneal white fat cells into brown-like adipocytes; and pRB is inactivated by phosphorylation during cold-induced activation and recruitment of BAT [39]. However, definite proof that pRB differentially regulates the formation of white and brown fat cells in vivo has remained elusive.
Scimè et al. [206] recently reported that white adipose tissue mass in p107−/− mice is substantially decreased, whereas the weight of the interscapular BAT depot is similar to that of wild-type mice. Adipocytes present in p107-deficient WAT contain smaller lipid droplets, more mitochondria and express highly elevated levels of UCP1 and PGC-1α. Using a novel FACS-based strategy for isolating preadipocytes, it was demonstrated that WAT depots of p107-deficient animals contain 30% more preadipocytes than WAT from wild-type animals, indicating that p107 positively regulates differentiation of white preadipocytes in vivo. In contrast, cultures of primary preadipocytes from adult p107 null mice have an increased differentiation potential in vitro, and the resulting p107−/− fat cells express UCP1 and PGC-1α. The finding that freshly isolated primary preadipocytes from inguinal WAT of adult p107−/− mice contain dramatically decreased levels of pRB furthermore suggests that pRB may in fact mediate the effect of p107-deficiency on adipose conversion [206].
It is conceivable that pRB controls white versus brown adipogenesis by interfering with expression of PGC-1α. In comparison with wild-type cells, Rb−/− MEFs exhibit increased cAMP sensitivity during the early stages of differentiation. Increased cAMP sensitivity is important for the brown adipogenic fate of fibroblasts lacking pRB, as an inhibitor of PKA attenuates the induction of PGC-1α and Tfam expression and largely blocks induction of UCP1 expression [39]. Scimè et al. [206] applied ChIP to show pRB association with at least two regions of the PGC-1α promoter, located approx. 1 and 4 kb upstream of the transcription start site. Moreover, pRB represses the activity of the PGC-1α promoter in a dose-dependent manner, a repressive effect mimicked by E2F4 expression and amplified by co-expression of pRB and E2F4.
In summary, pRB and p107 are potent regulators of white versus brown adipose conversion in vitro and in vivo. However, it remains to be shown how pRB regulates cAMP sensitivity, how pRB is recruited to the PGC-1α promoter in preadipocytes and whether the repressive effect of pRB is dependent on histone deacetylase activity. Finally, it is not clear why p107-deficient preadipocytes are impaired in differentiation in vivo, but display improved differentiation in vitro.
Wnt signalling
Activation of Wnt signalling potently inhibits white adipocyte differentiation in vitro and in vivo [207,208]. Activated Wnt signalling prevents induction of C/EBPα and PPARγ during preadipocyte differentiation, and forced expression of these overrules the differentiation-inhibitory effects of Wnt signalling [207]. The mechanism by which Wnt signalling blocks induction of C/EBPα and PPARγ expression is unknown. Wnt signalling also blocks differentiation of brown preadipocytes [16,209]. Although ectopic expression of C/EBPα and PPARγ allows brown preadipocytes to accumulate lipid droplets in the presence of activated Wnt signalling, they do not express typical brown adipocyte genes, including PGC-1α and UCP1. Combined ectopic expression of PPARγ and PGC-1α, however, overrules the effect of Wnt signalling on both differentiation and expression of brown adipocyte genes [16]. Interestingly, activation of Wnt signalling in immature brown adipocytes shuts down brown adipocyte genes, while leaving general adipocyte transcription factors and marker genes untouched [16]. The selective down-regulation of brown adipocyte genes by Wnt signalling in this scenario is likely to be due to the observed rapid attenuation of PGC-1α expression, as forced expression of PGC-1α bypasses the effect of activated Wnt signalling. It is not known how activated Wnt signalling leads to down-regulation of PGC-1α expression. Wnt signalling also affects BAT in vivo. FABP4–Wnt10b transgenic mice have dysfunctional interscapular adipose tissue, with adipocytes displaying a unilocular morphology [208]. Moreover, these mice are cold-sensitive and fail to increase core body temperature in response to injection with a β-adrenergic agonist [16,208]. Similarly, UCP1–Wnt10b transgenic mice have impaired BAT function, with interscapular adipocytes exhibiting unilocular lipid droplets and strongly reduced expression of brown adipocyte genes [16]. Finally, Wnt10b is up-regulated in BAT from genetically obese mice, concomitantly with a down-regulation of PGC-1α and UCP1 expression [16], suggesting that Wnt signalling may also affect brown adipocyte function in a physiological setting.
SIRT3 (sirtuin 3)
SIRT3 is one of seven members of the mammalian sirtuin family possessing NAD+-dependent protein deacetylase and ADP-ribosylase activities [210]. Mouse SIRT3 is ubiquitously expressed, but displays an enriched expression in BAT relative to WAT [15]. Moreover, it localizes to the mitochondrial inner membrane. Forced expression of wild-type SIRT3 in HIB-1B cells enhances expression of UCP1, PGC-1α and enzymes of the respiratory chain, whereas expression of a dominant-negative SIRT3 attenuates expression of these genes [15]. The effect of the dominant-negative SIRT3 is partially bypassed by ectopic expression of PGC-1α, suggesting that SIRT3 promotes brown adipogenesis at least in part through induction of PGC-1α expression. The induction of PGC-1α expression is possibly linked to an increased activity of CREB, as the level of phosphorylated CREB was enhanced in SIRT3-expressing HIB-1B cells [15]. Physiologically, the expression of SIRT3 is induced in BAT in response to cold exposure, whereas the expression of SIRT3 is down-regulated in BAT of genetically obese mice [15]. It is noteworthy that another sirtuin family member, namely SIRT1, has been shown to inhibit adipocyte differentiation of 3T3-L1 preadipocytes and to promote triacylglycerol mobilization in 3T3-L1 adipocytes [211]. SIRT1 mediated these effects by promoting the recruitment of co-repressors to PPARγ, thereby blunting PPARγ activity [211]. Ectopic expression of SIRT1 in HIB-1B cells has, however, no effect on either adipose conversion or expression of PGC-1α and UCP1 [15].
NO (nitric oxide)
NO is a physiological regulator of a plethora of tasks, many of which involve its effects on mitochondria [212]. NO has been reported to stimulate the differentiation of primary rat brown preadipocytes [213]. Subsequently, NO was shown to induce mitochondrial biogenesis in brown adipocytes, and this effect of NO was dependent on an NO-mediated induction of PGC-1α expression [214]. Adipocytes appear to express eNOS (endothelial NO synthase) as the only NOS [214]. eNOS knockout mice have impaired BAT function with an increased presence of unilocular adipocytes, a decreased mitochondrial content and a decreased expression of PGC-1α and UCP1. Consistently, eNOS-deficient mice were sensitive to diet-induced weight gain [214]. Although eNOS and NO are clearly capable of inducing mitochondrial biogenesis in brown adipocytes, the effect is not restricted to this cell type. NO induces mitochondrial biogenesis also in 3T3-L1 adipocytes and in several non-adipose cell lines, and eNOS−/− mice exhibit reduced amounts of mitochondria in all tissues examined, with WAT being the sole exception [214].
CONCLUSIONS, FUTURE DIRECTIONS AND PERSPECTIVES
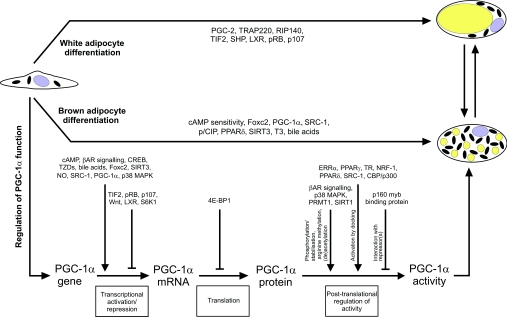
The ability of brown fat to dissipate energy through uncoupled respiration and thermogenesis has caused speculation as to whether controlled recruitment of brown adipocytes would be a suitable strategy to counteract obesity. Most crucial regulators of adipogenesis have possibly been identified, like PPARγ and C/EBPs. However, novel important transcription factors involved in white adipose conversion have recently been reported [215,216], and several factors affecting both white and brown adipogenesis have been described, such as components of the Wnt signalling pathway [16,207] and GATA transcription factors [217,218]. Knowledge of factors specifically involved in promoting or inhibiting brown adipocyte differentiation is sparse. However, research in recent years have brought to light some examples of potential regulators. With few exceptions, the recurrent impact of these is direct or indirect effects on the activity of PGC-1α (Figure 1). The ability of PGC-1α to induce expression of UCP1 as well as mitochondrial biogenesis and activity in fat cells points to PGC-1α as the prime factor co-ordinating brown adipocyte differentiation and function. The activity of PGC-1α is controlled by the potential regulators at multiple levels: through positive or negative effects on transcription of the PGC-1α gene, through translational regulation of the PGC-1α mRNA or through post-translational modification of PGC-1α. The post-translational regulation of PGC-1α includes interactions with repressor proteins, p38 MAPK-dependent phosphorylation of PGC-1α resulting in its activation and stabilization (and the dissociation of repressors), and recruitment of PGC-1α to target promoters through interaction with transcription factors, leading to activation of PGC-1α due to the docking effect (Figure 1). In several cases, the molecular mechanism through which the factors discussed above modulate PGC-1α activity is unknown, e.g. how does S6K1 and Wnt signalling decrease expression of PGC-1α, and does the improved translation of the PGC-1α mRNA in the absence of 4E-BP1 occur as a result of a direct effect of 4E-BP1 on the PGC-1α mRNA?
Figure 1. Molecular determinants involved in white versus brown adipocyte differentiation.
Factors that characterize white or brown adipogenesis are indicated in the upper and middle parts of the Figure respectively, whereas factors influencing PGC-1α activity at various levels are listed in the lower part. The mechanism by which LXR affects UCP1 expression in fat and white versus brown adipose conversion is unknown. The potential transdifferentiation of white and brown fat cells is indicated in the upper right part of the Figure. See the text for details. The involvement of TRAP220 and PGC-2 in white adipose conversion has been described [219,220].
Of all the potential regulators, only three examples have been reported in which the expression of PGC-1α mRNA was not affected, and in one example, although not addressed, no change in PGC-1α expression is suspected. The latter example is the Cidea knockout mice, where Cidea has been proposed to act through a direct interaction with the UCP1 protein and thus downstream of PGC-1α. The former three examples are the transgenic mice expressing a constitutively active PPARδ in fat, the RIP140-deficient mice and the LXRβ−/− and LXRα−/−/LXRβ−/− knockout mice. As PPARδ interacts with PGC-1α and effectively is co-activated by PGC-1α, it is plausible that PPARδ activates PGC-1α through the transcription factor docking mechanism. In this way, PPARδ could increase PGC-1α activity without affecting its expression. In the case of RIP140 knockout mice it appears that the absence of RIP140 allows ectopic activation of the UCP1 promoter by low levels of PGC-1α in WAT. On the basis of the correlation between increased activity of PGC-1α and hyperactive BAT as well as brown adipose-like conversion of WAT, it is also possible that RIP140 represses PGC-1α activity through an unknown mechanism. Several possibilities exist: RIP140 might repress PGC-1α activity through a direct interaction between the two or RIP140 might sequester PGC-1α-interacting proteins necessary for execution of PGC-1α-mediated functions. In the LXRβ−/− single knockout and LXRα−/−/LXRβ−/− double knockout mice it remains to be determined if the level of the PGC-1α protein is increased.
Whether controlled recruitment of brown fat cells is a suitable strategy in the treatment of obesity and obesity-associated disorders remains to be shown. Nevertheless, designed interference of the balance between white and brown adipocyte differentiation requires a more detailed understanding of factors differentially regulating the formation of white and brown fat cells. In addition, and perhaps even more important in relation to humans, would be detailed knowledge of the processes that control the regression of BAT and/or conversion of BAT into largely non-adrenergic responsive WAT after birth in larger mammals. A few regulators of white versus brown fat cell differentiation have been described, and from the reported data, it appears that most of these act upstream of PGC-1α. However, the existence of factors that promote brown adipose conversion in a PGC-1α-independent manner cannot be ruled out. Moreover, a significant impact of other PGC-1 family members is highly likely. Being an active area of research, the coming years will undoubtedly expose novel and exciting insight into regulatory circuits controlling white versus brown adipocyte differentiation.
Acknowledgments
Work in our laboratories is supported by the Carlsberg Foundation (grant to J. B. H.), the Danish Health Science Research Council (grant to J. B. H.), the Augustinus Foundation (grant to J. B. H.), the Novo Nordisk Foundation (grants to J. B. H. and K. K.), the Danish Biotechnology Program (grant to K. K.), the Danish Cancer Society (grant to K. K.), the Danish Natural Science Research Council (grant to K. K.) and the European Community (grant QLK1-CT-2001-00183 to K. K.).
References
- 1.Cinti S. The adipose organ. Prostaglandins Leukotrienes Essent. Fatty Acids. 2005;73:9–15. doi: 10.1016/j.plefa.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 3.Himms-Hagen J. Does brown adipose tissue (BAT) have a role in the physiology or treatment of human obesity? Rev. Endocr. Metab. Disord. 2001;2:395–401. doi: 10.1023/a:1011856617047. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Barroso M. D., Ricquier D., Cassard-Doulcier A. M. The human uncoupling protein-1 gene (UCP1): present status and perspectives in obesity research. Obes. Rev. 2000;1:61–72. doi: 10.1046/j.1467-789x.2000.00009.x. [DOI] [PubMed] [Google Scholar]
- 5.Collins S., Cao W., Robidoux J. Learning new tricks from old dogs: β-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol. Endocrinol. 2004;18:2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- 6.Dulloo A. G., Miller D. S. Energy balance following sympathetic denervation of brown adipose tissue. Can. J. Physiol. Pharmacol. 1984;62:235–240. doi: 10.1139/y84-035. [DOI] [PubMed] [Google Scholar]
- 7.Hamann A., Flier J. S., Lowell B. B. Decreased brown fat markedly enhances susceptibility to diet-induced obesity, diabetes, and hyperlipidemia. Endocrinology. 1996;137:21–29. doi: 10.1210/endo.137.1.8536614. [DOI] [PubMed] [Google Scholar]
- 8.Lowell B. B., Susulic V., Hamann A., Lawitts J. A., Himms-Hagen J., Boyer B. B., Kozak L. P., Flier J. S. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature (London) 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 9.Melnyk A., Harper M. E., Himms-Hagen J. Raising at thermoneutrality prevents obesity and hyperphagia in BAT-ablated transgenic mice. Am. J. Physiol. 1997;41:R1088–R1093. doi: 10.1152/ajpregu.1997.272.4.R1088. [DOI] [PubMed] [Google Scholar]
- 10.Enerbäck S., Jacobsson A., Simpson E. M., Guerra C., Yamashita H., Harper M., Kozak L. P. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature (London) 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 11.Kopecky J., Hodny Z., Rossmeisl M., Syrovy I., Kozak L. P. Reduction of dietary obesity in aP2-Ucp transgenic mice: physiology and adipose tissue distribution. Am. J. Physiol. Endocrinol. Metab. 1996;270:E768–E775. doi: 10.1152/ajpendo.1996.270.5.E768. [DOI] [PubMed] [Google Scholar]
- 12.Kopecky J., Clarke G., Enerback S., Spiegelman B., Kozak L. P. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J. Clin. Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefl B., Janovska A., Hodny Z., Rossmeisl M., Horakova M., Syrovy I., Bemova J., Bendlova B., Kopecky J. Brown fat is essential for cold-induced thermogenesis but not for obesity resistance in aP2-Ucp mice. Am. J. Physiol. 1998;37:E527–E533. doi: 10.1152/ajpendo.1998.274.3.E527. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z., Yon Toh S., Chen Z., Guo K., Ng C. P., Ponniah S., Lin S. C., Hong W., Li P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat. Genet. 2003;35:49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
- 15.Shi T., Wang F., Stieren E., Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 16.Kang S., Bajnok L., Longo K. A., Petersen R. K., Hansen J. B., Kristiansen K., MacDougald O. A. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1α. Mol. Cell. Biol. 2005;25:1272–1282. doi: 10.1128/MCB.25.4.1272-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moulin K., Truel N., Andre M., Arnauld E., Nibbelink M., Cousin B., Dani C., Penicaud L., Casteilla L. Emergence during development of the white-adipocyte cell phenotype is independent of the brown-adipocyte cell phenotype. Biochem. J. 2001;356:659–664. doi: 10.1042/0264-6021:3560659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaus S. Functional differentiation of white and brown adipocytes. BioEssays. 1997;19:215–223. doi: 10.1002/bies.950190307. [DOI] [PubMed] [Google Scholar]
- 19.Loncar D., Afzelius B. A. Ontogenetical changes in adipose tissue of the cat: convertible adipose tissue. J. Ultrastruct. Mol. Struct. Res. 1989;102:9–23. doi: 10.1016/0889-1605(89)90028-1. [DOI] [PubMed] [Google Scholar]
- 20.Ashwell M., Stirling D., Freeman S., Holloway B. R. Immunological, histological and biochemical assessment of brown adipose tissue activity in neonatal, control and β-stimulant-treated adult dogs. Int. J. Obes. 1987;11:357–365. [PubMed] [Google Scholar]
- 21.Trayhurn P., Thomas M. E., Keith J. S. Postnatal development of uncoupling protein, uncoupling protein mRNA, and Glut4 in adipose tissues of goats. Am. J. Physiol. 1993;265:R676–R682. doi: 10.1152/ajpregu.1993.265.3.R676. [DOI] [PubMed] [Google Scholar]
- 22.Casteilla L., Champigny O., Bouillaud F., Robelin J., Ricquier D. Sequential changes in the expression of mitochondrial protein mRNA during the development of brown adipose tissue in bovine and ovine species. Biochem. J. 1989;257:665–671. doi: 10.1042/bj2570665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soppela P., Nieminen M., Saarela S., Keith J. S., Morrison J. N., Macfarlane F., Trayhurn P. Brown fat-specific mitochondrial uncoupling protein in adipose tissues of newborn reindeer. Am. J. Physiol. 1991;260:R1229–R1234. doi: 10.1152/ajpregu.1991.260.6.R1229. [DOI] [PubMed] [Google Scholar]
- 24.Lean M. E., James W. P., Jennings G., Trayhurn P. Brown adipose tissue uncoupling protein content in human infants, children and adults. Clin. Sci. 1986;71:291–297. doi: 10.1042/cs0710291. [DOI] [PubMed] [Google Scholar]
- 25.Loncar D., Bedrica L., Mayer J., Cannon B., Nedergaard J., Afzelius B. A., Svajger A. The effect of intermittent cold treatment on the adipose tissue of the cat: apparent transformation from white to brown adipose tissue. J. Ultrastruct. Mol. Struct. Res. 1986;97:119–129. doi: 10.1016/s0889-1605(86)80012-x. [DOI] [PubMed] [Google Scholar]
- 26.Champigny O., Ricquier D., Blondel O., Mayers R. M., Briscoe M. G., Holloway B. R. β-3-Adrenergic receptor stimulation restores message and expression of brown-fat mitochondrial uncoupling protein in adult dogs. Proc. Natl. Acad. Sci. U.S.A. 1991;88:10774–10777. doi: 10.1073/pnas.88.23.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nougues J., Reyne Y., Champigny O., Holloway B., Casteilla L., Ricquier D. The β-3-adrenoceptor agonist ICI-D7114 is not as efficient on reinduction of uncoupling protein mRNA in sheep as it is in dogs and smaller species. J. Anim. Sci. 1993;71:2388–2394. doi: 10.2527/1993.7192388x. [DOI] [PubMed] [Google Scholar]
- 28.Oberkofler H., Dallinger G., Liu Y. M., Hell E., Krempler F., Patsch W. Uncoupling protein gene: quantification of expression levels in adipose tissues of obese and non-obese humans. J. Lipid Res. 1997;38:2125–2133. [PubMed] [Google Scholar]
- 29.Huttunen P., Hirvonen J., Kinnula V. The occurrence of brown adipose tissue in outdoor workers. Eur. J. Appl. Physiol. Occup. Physiol. 1981;46:339–345. doi: 10.1007/BF00422121. [DOI] [PubMed] [Google Scholar]
- 30.Leppaluoto J., Paakkonen T., Korhonen I., Hassi J. Pituitary and autonomic responses to cold exposures in man. Acta Physiol. Scand. 2005;184:255–264. doi: 10.1111/j.1365-201X.2005.01464.x. [DOI] [PubMed] [Google Scholar]
- 31.Ricquier D., Nechad M., Mory G. Ultrastructural and biochemical characterization of human brown adipose tissue in pheochromocytoma. J. Clin. Endocrinol. Metab. 1982;54:803–807. doi: 10.1210/jcem-54-4-803. [DOI] [PubMed] [Google Scholar]
- 32.Lean M. E., James W. P., Jennings G., Trayhurn P. Brown adipose tissue in patients with pheochromocytoma. Int. J. Obes. 1986;10:219–227. [PubMed] [Google Scholar]
- 33.Zancanaro C., Pelosi G., Accordini C., Balercia G., Sbabo L., Cinti S. Immunohistochemical identification of the uncoupling protein in human hibernoma. Biol. Cell. 1994;80:75–78. doi: 10.1016/0248-4900(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 34.Cousin B., Cinti S., Morroni M., Raimbault S., Ricquier D., Penicaud L., Casteilla L. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J. Cell Sci. 1992;103:931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- 35.Loncar D. Convertible adipose tissue in mice. Cell Tissue Res. 1991;266:149–161. doi: 10.1007/BF00678721. [DOI] [PubMed] [Google Scholar]
- 36.Guerra C., Koza R. A., Yamashita H., Walsh K., Kozak L. P. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Invest. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Himms-Hagen J., Melnyk A., Zingaretti M. C., Ceresi E., Barbatelli G., Cinti S. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol. 2000;279:C670–C681. doi: 10.1152/ajpcell.2000.279.3.C670. [DOI] [PubMed] [Google Scholar]
- 38.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 39.Hansen J. B., Jørgensen C., Petersen R. K., Hallenborg P., De Matteis R., Bøye H. A., Petrovic N., Enerbäck S., Nedergaard J., Cinti S., et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiraby C., Tavernier G., Lefort C., Larrouy D., Bouillaud F., Ricquier D., Langin D. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- 41.Bachman E. S., Dhillon H., Zhang C. y., Cinti S., Bianco A. C., Kobilka B. K., Lowell B. B. βAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–845. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 42.Gorla-Bajszczak A., Siegrist-Kaiser C., Boss O., Burger A. G., Meier C. A. Expression of peroxisome proliferator-activated receptors in lean and obese Zucker rats. Eur. J. Endocrinol. 2000;142:71–78. doi: 10.1530/eje.0.1420071. [DOI] [PubMed] [Google Scholar]
- 43.Lindgren E. M., Nielsen R., Petrovic N., Jacobsson A., Mandrup S., Cannon B., Nedergaard J. Noradrenaline represses PPAR (peroxisome-proliferator-activated receptor) γ2 gene expression in brown adipocytes: intracellular signalling and effects on PPARγ2 and PPARγ1 protein levels. Biochem. J. 2004;382:597–606. doi: 10.1042/BJ20031622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bengtsson T., Cannon B., Nedergaard J. Differential adrenergic regulation of the gene expression of the β-adrenoceptor subtypes β1, β2 and β3 in brown adipocytes. Biochem. J. 2000;347:643–651. [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang C. S., Loftus T. M., Mandrup S., Lane M. D. Adipocyte differentiation and leptin expression. Annu. Rev. Cell Dev. Biol. 1997;13:231–259. doi: 10.1146/annurev.cellbio.13.1.231. [DOI] [PubMed] [Google Scholar]
- 46.Klein J., Fasshauer M., Klein H. H., Benito M., Kahn C. R. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. BioEssays. 2002;24:382–388. doi: 10.1002/bies.10058. [DOI] [PubMed] [Google Scholar]
- 47.Ross S. R., Choy L., Graves R. A., Fox N., Solevjeva V., Klaus S., Ricquier D., Spiegelman B. M. Hibernoma formation in transgenic mice and isolation of a brown adipocyte cell line expressing the uncoupling protein gene. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7561–7565. doi: 10.1073/pnas.89.16.7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benito M., Porras A., Santos E. Establishment of permanent brown adipocyte cell lines achieved by transfection with SV40 large T antigen and ras genes. Exp. Cell Res. 1993;209:248–254. doi: 10.1006/excr.1993.1308. [DOI] [PubMed] [Google Scholar]
- 49.Kozak U. C., Kozak L. P. Norepinephrine-dependent selection of brown adipocyte cell lines. Endocrinology. 1994;134:906–913. doi: 10.1210/endo.134.2.7905411. [DOI] [PubMed] [Google Scholar]
- 50.Irie Y., Asano A., Canas X., Nikami H., Aizawa S., Irie Y., Asano A., Saito M. Immortal brown adipocytes from p53-knockout mice: differentiation and expression of uncoupling proteins. Biochem. Biophys. Res. Commun. 1999;255:221–225. doi: 10.1006/bbrc.1998.9999. [DOI] [PubMed] [Google Scholar]
- 51.Klein J., Fasshauer M., Ito M., Lowell B. B., Benito M., Kahn C. R. β3-Adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in brown adipocytes. J. Biol. Chem. 1999;274:34795–34802. doi: 10.1074/jbc.274.49.34795. [DOI] [PubMed] [Google Scholar]
- 52.Soukas A., Socci N. D., Saatkamp B. D., Novelli S., Friedman J. M. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J. Biol. Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- 53.Rohlfs E. M., Daniel K. W., Premont R. T., Kozak L. P., Collins S. Regulation of the uncoupling protein gene (Ucp) by β1, β2, and β3-adrenergic receptor subtypes in immortalized brown adipose cell lines. J. Biol. Chem. 1995;270:10723–10732. doi: 10.1074/jbc.270.18.10723. [DOI] [PubMed] [Google Scholar]
- 54.Valmaseda A., Carmona M. C., Barbera M. J., Vinas O., Mampel T., Iglesias R., Villarroya F., Giralt M. Opposite regulation of PPAR-α and -γ gene expression by both their ligands and retinoic acid in brown adipocytes. Mol. Cell. Endocrinol. 1999;154:101–109. doi: 10.1016/s0303-7207(99)00081-7. [DOI] [PubMed] [Google Scholar]
- 55.Rosen E. D., Walkey C. J., Puigserver P., Spiegelman B. M. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 56.Unami A., Shinohara Y., Kajimoto K., Baba Y. Comparison of gene expression profiles between white and brown adipose tissues of rat by microarray analysis. Biochem. Pharmacol. 2004;67:555–564. doi: 10.1016/j.bcp.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 57.Issemann I., Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 58.Leonard J. L., Mellen S. A., Larsen P. R. Thyroxine 5′-deiodinase activity in brown adipose tissue. Endocrinology. 1983;112:1153–1155. doi: 10.1210/endo-112-3-1153. [DOI] [PubMed] [Google Scholar]
- 59.Maffei M., Fei H., Lee G., Dani C., Leroy P., Zhang Y., Proenca R., Negrel R., Ailhaud G., Friedman J. M. Increased expression in adipocytes of ob RNA in mice with lesions of the hypothalamus and with mutations at the db locus. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6957–6960. doi: 10.1073/pnas.92.15.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boeuf S., Klingenspor M., Van Hal N. L., Schneider T., Keijer J., Klaus S. Differential gene expression in white and brown preadipocytes. Physiol. Genomics. 2001;7:15–25. doi: 10.1152/physiolgenomics.00048.2001. [DOI] [PubMed] [Google Scholar]
- 61.Jimenez M., Leger B., Canola K., Lehr L., Arboit P., Seydoux J., Russell A. P., Giacobino J. P., Muzzin P., Preitner F. β1/β2/β3-Adrenoceptor knockout mice are obese and cold-sensitive but have normal lipolytic responses to fasting. FEBS Lett. 2002;530:37–40. doi: 10.1016/s0014-5793(02)03387-2. [DOI] [PubMed] [Google Scholar]
- 62.Chiu C. H., Lin W. D., Huang S. Y., Lee Y. H. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004;18:1970–1975. doi: 10.1101/gad.1213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cummings D. E., Brandon E. P., Planas J. V., Motamed K., Idzerda R. L., McKnight G. S. Genetically lean mice result from targeted disruption of the RIIβ subunit of protein kinase A. Nature (London) 1996;382:622–626. doi: 10.1038/382622a0. [DOI] [PubMed] [Google Scholar]
- 64.Cederberg A., Gronning L. M., Ahren B., Tasken K., Carlsson P., Enerbäck S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 65.Gronning L. M., Cederberg A., Miura N., Enerbäck S., Tasken K. Insulin and TNFα induce expression of the forkhead transcription factor gene Foxc2 in 3T3-L1 adipocytes via PI3K and ERK1/2-dependent pathways. Mol. Endocrinol. 2002;16:873–883. doi: 10.1210/mend.16.4.0803. [DOI] [PubMed] [Google Scholar]
- 66.Leonardsson G., Steel J. H., Christian M., Pocock V., Milligan S., Bell J., So P. W., Medina-Gomez G., Vidal-Puig A., White R., Parker M. G. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis K. E., Moldes M., Farmer S. R. The forkhead transcription factor FoxC2 inhibits white adipocyte differentiation. J. Biol. Chem. 2004;279:42453–42461. doi: 10.1074/jbc.M402197200. [DOI] [PubMed] [Google Scholar]
- 68.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 69.Kelly D. P., Scarpulla R. C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 70.Mootha V. K., Handschin C., Arlow D., Xie X., Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Willy P. J., Schulman I. G., Heyman R. A., Lander E. S., Spiegelman B. M. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schreiber S. N., Emter R., Hock M. B., Knutti D., Cardenas J., Podvinec M., Oakeley E. J., Kralli A. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B. M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 73.Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature (London) 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 74.Handschin C., Rhee J., Lin J., Tarr P. T., Spiegelman B. M. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin J., Wu P. H., Tarr P. T., Lindenberg K. S., St-Pierre J., Zhang C. Y., Mootha V. K., Jager S., Vianna C. R., Reznick R. M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 76.Leone T. C., Lehman J. J., Finck B. N., Schaeffer P. J., Wende A. R., Boudina S., Courtois M., Wozniak D. F., Sambandam N., Bernal-Mizrachi C., et al. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:672–687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin J., Handschin C., Spiegelman B. M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 78.Puigserver P., Rhee J., Lin J., Wu Z., Yoon J. C., Zhang C. Y., Krauss S., Mootha V. K., Lowell B. B., Spiegelman B. M. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 79.Cao W., Daniel K. W., Robidoux J., Puigserver P., Medvedev A. V., Bai X., Floering L. M., Spiegelman B. M., Collins S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 2004;24:3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao W., Medvedev A. V., Daniel K. W., Collins S. β-Adrenergic activation of p38 MAP kinase in adipocytes. J. Biol. Chem. 2001;276:27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 81.Puigserver P., Adelmant G., Wu Z., Fan M., Xu J., O'Malley B., Spiegelman B. M. Activation of PPAR coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 82.Fan M., Rhee J., St-Pierre J., Handschin C., Puigserver P., Lin J., Jaeger S., Erdjument-Bromage H., Tempst P., Spiegelman B. M. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knutti D., Kressler D., Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ichida M., Nemoto S., Finkel T. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptorγcoactivator-1α (PGC-1α) J. Biol. Chem. 2002;277:50991–50995. doi: 10.1074/jbc.M210262200. [DOI] [PubMed] [Google Scholar]
- 85.Teyssier C., Ma H., Emter R., Kralli A., Stallcup M. R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nemoto S., Ferguson M. M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 87.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature (London) 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 88.Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B. M. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 89.Kressler D., Schreiber S. N., Knutti D., Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor α. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 90.Andersson U., Scarpulla R. C. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.St-Pierre J., Lin J., Krauss S., Tarr P. T., Yang R., Newgard C. B., Spiegelman B. M. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 92.Meirhaeghe A., Crowley V., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J. K., Yeo G., et al. Characterization of the human, mouse and rat PGC1β (peroxisome-proliferator-activated receptor-γ co-activator 1β) gene in vitro and in vivo. Biochem. J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kamei Y., Ohizumi H., Fujitani Y., Nemoto T., Tanaka T., Takahashi N., Kawada T., Miyoshi M., Ezaki O., Kakizuka A. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Savagner F., Mirebeau D., Jacques C., Guyetant S., Morgan C., Franc B., Reynier P., Malthiery Y. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem. Biophys. Res. Commun. 2003;310:779–784. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 95.Gleyzer N., Vercauteren K., Scarpulla R. C. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Picard F., Gehin M., Annicotte J. S., Rocchi S., Champy M. F., O'Malley B. W., Chambon P., Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 97.Spencer T. E., Jenster G., Burcin M. M., Allis C. D., Zhou J., Mizzen C. A., McKenna N. J., Onate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 98.Hoang T., Fenne I. S., Cook C., Borud B., Bakke M., Lien E. A., Mellgren G. cAMP-dependent protein kinase regulates ubiquitin–proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J. Biol. Chem. 2004;279:49120–49130. doi: 10.1074/jbc.M409746200. [DOI] [PubMed] [Google Scholar]
- 99.Wang Z., Qi C., Krones A., Woodring P., Zhu X., Reddy J. K., Evans R. M., Rosenfeld M. G., Hunter T. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab. 2006;3:111–122. doi: 10.1016/j.cmet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 100.Cavailles V., Dauvois S., L'Horset F., Lopez G., Hoare S., Kushner P. J., Parker M. G. Nuclear factor Rip140 modulates transcriptional activation by the estrogen-receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.L'Horset F., Dauvois S., Heery D. M., Cavailles V., Parker M. G. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol. Cell. Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Treuter E., Albrektsen T., Johansson L., Leers J., Gustafsson J. A. A regulatory role for RIP140 in nuclear receptor activation. Mol. Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 103.Steel J. H., White R., Parker M. G. Role of the RIP140 corepressor in ovulation and adipose biology. J. Endocrinol. 2005;185:1–9. doi: 10.1677/joe.1.05896. [DOI] [PubMed] [Google Scholar]
- 104.White R., Leonardsson G., Rosewell I., Ann Jacobs M., Milligan S., Parker M. The nuclear receptor co-repressor Nrip1 (RIP140) is essential for female fertility. Nat. Med. 2000;6:1368–1374. doi: 10.1038/82183. [DOI] [PubMed] [Google Scholar]
- 105.Christian M., Kiskinis E., Debevec D., Leonardsson G., White R., Parker M. G. RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 2005;25:9383–9391. doi: 10.1128/MCB.25.21.9383-9391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Powelka A. M., Seth A., Virbasius J. V., Kiskinis E., Nicoloro S. M., Guilherme A., Tang X., Straubhaar J., Cherniack A. D., Parker M. G., Czech M. P. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J. Clin. Invest. 2006;116:125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klaus S., Choy L., Champigny O., Cassard-Doulcier A. M., Ross S., Spiegelman B., Ricquier D. Characterization of the novel brown adipocyte cell line HIB 1B. Adrenergic pathways involved in regulation of uncoupling protein gene expression. J. Cell Sci. 1994;107:313–319. doi: 10.1242/jcs.107.1.313. [DOI] [PubMed] [Google Scholar]
- 108.Silva J. E. Thermogenic mechanisms and their hormonal regulation. Physiol. Rev. 2006;86:435–464. doi: 10.1152/physrev.00009.2005. [DOI] [PubMed] [Google Scholar]
- 109.Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature (London) 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 110.Xue B. Z., Coulter A., Rim J. S., Koza R. A., Kozak L. P. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Mol. Cell. Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Golozoubova V., Gullberg H., Matthias A., Cannon B., Vennstrom B., Nedergaard J. Depressed thermogenesis but competent brown adipose tissue recruitment in mice devoid of all hormone-binding thyroid hormone receptors. Mol. Endocrinol. 2004;18:384–401. doi: 10.1210/me.2003-0267. [DOI] [PubMed] [Google Scholar]
- 112.Rosen E. D., Hsu C. H., Wang X., Sakai S., Freeman M. W., Gonzalez F. J., Spiegelman B. M. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rangwala S. M., Lazar M. A. Transcriptional control of adipogenesis. Annu. Rev. Nutr. 2000;20:535–559. doi: 10.1146/annurev.nutr.20.1.535. [DOI] [PubMed] [Google Scholar]
- 114.Rosen E. D., Sarraf P., Troy A. E., Bradwin G., Moore K., Milstone D. S., Spiegelman B. M., Mortensen R. M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 115.Barak Y., Nelson M. C., Ong E. S., Jones Y. Z., Ruiz-Lozano P., Chien K. R., Koder A., Evans R. M. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 116.Jones J. R., Barrick C., Kim K. A., Lindner J., Blondeau B., Fujimoto Y., Shiota M., Kesterson R. A., Kahn B. B., Magnuson M. A. Deletion of PPARγ in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.He W., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J. M., Evans R. M. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koutnikova H., Cock T. A., Watanabe M., Houten S. M., Champy M. F., Dierich A., Auwerx J. Compensation by the muscle limits the metabolic consequences of lipodystrophy in PPARγ hypomorphic mice. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14457–14462. doi: 10.1073/pnas.2336090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang J., Fu M., Cui T., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Song Q., Chen Y. E. Selective disruption of PPARγ2 impairs the development of adipose tissue and insulin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10703–10708. doi: 10.1073/pnas.0403652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Imai T., Takakuwa R., Marchand S., Dentz E., Bornert J. M., Messaddeq N., Wendling O., Mark M., Desvergne B., Wahli W., et al. Peroxisome proliferator-activated receptor γ is required in mature white and brown adipocytes for their survival in the mouse. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4543–4547. doi: 10.1073/pnas.0400356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lehmann J. M., Moore L. B., Smith-Oliver T. A., Wilkison W. O., Willson T. M., Kliewer S. A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ. J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 122.Tai T. A., Jennermann C., Brown K. K., Oliver B. B., MacGinnitie M. A., Wilkison W. O., Brown H. R., Lehmann J. M., Kliewer S. A., Morris D. C., Graves R. A. Activation of the nuclear receptor peroxisome proliferator-activated receptor γ promotes brown adipocyte differentiation. J. Biol. Chem. 1996;271:29909–29914. doi: 10.1074/jbc.271.47.29909. [DOI] [PubMed] [Google Scholar]
- 123.Teruel T., Hernandez R., Benito M., Lorenzo M. Rosiglitazone and retinoic acid induce uncoupling protein-1 (UCP-1) in a p38 mitogen-activated protein kinase-dependent manner in fetal primary brown adipocytes. J. Biol. Chem. 2003;278:263–269. doi: 10.1074/jbc.M207200200. [DOI] [PubMed] [Google Scholar]
- 124.Foellmi-Adams L. A., Wyse B. M., Herron D., Nedergaard J., Kletzien R. F. Induction of uncoupling protein in brown adipose tissue: synergy between norepinephrine and pioglitazone, an insulin-sensitizing agent. Biochem. Pharmacol. 1996;52:693–701. doi: 10.1016/0006-2952(96)00345-0. [DOI] [PubMed] [Google Scholar]
- 125.Kelly L. J., Vicario P. P., Thompson G. M., Candelore M. R., Doebber T. W., Ventre J., Wu M. S., Meurer R., Forrest M. J., Conner M. W., et al. Peroxisome proliferator-activated receptors γ and α mediate in vivo regulation of uncoupling protein (UCP-1, UCP-2, UCP-3) gene expression. Endocrinology. 1998;139:4920–4927. doi: 10.1210/endo.139.12.6384. [DOI] [PubMed] [Google Scholar]
- 126.Sell H., Berger J. P., Samson P., Castriota G., Lalonde J., Deshaies Y., Richard D. Peroxisome proliferator-activated receptor γ agonism increases the capacity for sympathetically mediated thermogenesis in lean and ob/ob mice. Endocrinology. 2004;145:3925–3934. doi: 10.1210/en.2004-0321. [DOI] [PubMed] [Google Scholar]
- 127.Fukui Y., Masui S., Osada S., Umesono K., Motojima K. A new thiazolidinedione, NC-2100, which is a weak PPARγ activator, exhibits potent antidiabetic effects and induces uncoupling protein 1 in white adipose tissue of KKAy obese mice. Diabetes. 2000;49:759–767. doi: 10.2337/diabetes.49.5.759. [DOI] [PubMed] [Google Scholar]
- 128.Wilson-Fritch L., Nicoloro S., Chouinard M., Lazar M. A., Chui P. C., Leszyk J., Straubhaar J., Czech M. P., Corvera S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wilson-Fritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guan H. P., Ishizuka T., Chui P. C., Lehrke M., Lazar M. A. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes Dev. 2005;19:453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hondares E., Mora O., Yubero P., de la Concepcion M. R., Iglesias R., Giralt M., Villarroya F. Thiazolidinediones and rexinoids induce peroxisome proliferator-activated receptor coactivator (PGC)-1α gene transcription: an autoregulatory loop controls PGC-1α expression in adipocytes via peroxisome proliferator-activated receptor γ coactivation. Endocrinology. 2006;147:2829–2838. doi: 10.1210/en.2006-0070. [DOI] [PubMed] [Google Scholar]
- 132.Smith C. L., O'Malley B. W. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr. Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 133.Rocchi S., Picard F., Vamecq J., Gelman L., Potier N., Zeyer D., Dubuquoy L., Bac P., Champy M. F., Plunket K. D. A unique PPARγ ligand with potent insulin-sensitizing yet weak adipogenic activity. Mol. Cell. 2001;8:737–747. doi: 10.1016/s1097-2765(01)00353-7. [DOI] [PubMed] [Google Scholar]
- 134.Rieusset J., Touri F., Michalik L., Escher P., Desvergne B., Niesor E., Wahli W. A new selective peroxisome proliferator-activated receptor γ antagonist with antiobesity and antidiabetic activity. Mol. Endocrinol. 2002;16:2628–2644. doi: 10.1210/me.2002-0036. [DOI] [PubMed] [Google Scholar]
- 135.Berger J. P., Petro A. E., Macnaul K. L., Kelly L. J., Zhang B. B., Richards K., Elbrecht A., Johnson B. A., Zhou G., Doebber T. W., et al. Distinct properties and advantages of a novel peroxisome proliferator-activated protein γ selective modulator. Mol. Endocrinol. 2003;17:662–676. doi: 10.1210/me.2002-0217. [DOI] [PubMed] [Google Scholar]
- 136.Misra P., Chakrabarti R., Vikramadithyan R. K., Bolusu G., Juluri S., Hiriyan J., Gershome C., Rajjak A., Kashireddy P., Yu S., et al. PAT5A: a partial agonist of peroxisome proliferator-activated receptor γ is a potent antidiabetic thiazolidinedione yet weakly adipogenic. J. Pharmacol. Exp. Ther. 2003;306:763–771. doi: 10.1124/jpet.103.049791. [DOI] [PubMed] [Google Scholar]
- 137.Teruel T., Clapham J. C., Smith S. A. PPARα activation by Wy 14643 induces transactivation of the rat UCP-1 promoter without increasing UCP-1 mRNA levels and attenuates PPARγ-mediated increases in UCP-1 mRNA levels induced by rosiglitazone in fetal rat brown adipocytes. Biochem. Biophys. Res. Commun. 1999;264:311–315. doi: 10.1006/bbrc.1999.1526. [DOI] [PubMed] [Google Scholar]
- 138.Barbera M. J., Schluter A., Pedraza N., Iglesias R., Villarroya F., Giralt M. Peroxisome proliferator-activated receptor α activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 2001;276:1486–1493. doi: 10.1074/jbc.M006246200. [DOI] [PubMed] [Google Scholar]
- 139.Oberkofler H., Esterbauer H., Linnemayr V., Strosberg A. D., Krempler F., Patsch W. Peroxisome proliferator-activated receptor (PPAR) γ coactivator-1 recruitment regulates PPAR subtype specificity. J. Biol. Chem. 2002;277:16750–16757. doi: 10.1074/jbc.M200475200. [DOI] [PubMed] [Google Scholar]
- 140.Tong Y., Hara A., Komatsu M., Tanaka N., Kamijo Y., Gonzalez F. J., Aoyama T. Suppression of expression of muscle-associated proteins by PPARα in brown adipose tissue. Biochem. Biophys. Res. Commun. 2005;336:76–83. doi: 10.1016/j.bbrc.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 141.Teruel T., Smith S. A., Peterson J., Clapham J. C. Synergistic activation of UCP-3 expression in cultured fetal rat brown adipocytes by PPARα and PPARγ ligands. Biochem. Biophys. Res. Commun. 2000;273:560–564. doi: 10.1006/bbrc.2000.2982. [DOI] [PubMed] [Google Scholar]
- 142.Goetzman E. S., Tian L., Wood P. A. Differential induction of genes in liver and brown adipose tissue regulated by peroxisome proliferator-activated receptor α during fasting and cold exposure in acyl-CoA dehydrogenase-deficient mice. Mol. Genet. Metab. 2005;84:39–47. doi: 10.1016/j.ymgme.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 143.Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J. Clin. Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tanaka T., Yamamoto J., Iwasaki S., Asaba H., Hamura H., Ikeda Y., Watanabe M., Magoori K., Ioka R. X., Tachibana K., et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Luquet S., Lopez-Soriano J., Holst D., Fredenrich A., Melki J., Rassoulzadegan M., Grimaldi P. A. Peroxisome proliferator-activated receptor δ controls muscle development and oxydative capability. FASEB J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 146.Wang Y. X., Zhang C. L., Yu R. T., Cho H. K., Nelson M. C., Bayuga-Ocampo C. R., Ham J., Kang H., Evans R. M. Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol. 2004;2:1532–1539. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brun R. P., Tontonoz P., Forman B. M., Ellis R., Chen J., Evans R. M., Spiegelman B. M. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 1996;10:974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 148.Jehl-Pietri C., Bastie C., Gillot I., Luquet S., Grimaldi P. A. Peroxisome-proliferator-activated receptor δ mediates the effects of long-chain fatty acids on post-confluent cell proliferation. Biochem. J. 2000;350:93–98. [PMC free article] [PubMed] [Google Scholar]
- 149.Hansen J. B., Zhang H., Rasmussen T. H., Petersen R. K., Flindt E. N., Kristiansen K. Peroxisome proliferator-activated receptor δ (PPARδ)-mediated regulation of preadipocyte proliferation and gene expression is dependent on cAMP signaling. J. Biol. Chem. 2001;276:3175–3182. doi: 10.1074/jbc.M005567200. [DOI] [PubMed] [Google Scholar]
- 150.Bastie C., Holst D., Gaillard D., Jehl-Pietri C., Grimaldi P. A. Expression of peroxisome proliferator-activated receptor PPARδ promotes induction of PPARγ and adipocyte differentiation in 3T3C2 fibroblasts. J. Biol. Chem. 1999;274:21920–21925. doi: 10.1074/jbc.274.31.21920. [DOI] [PubMed] [Google Scholar]
- 151.Bastie C., Luquet S., Holst D., Jehl-Pietri C., Grimaldi P. A. Alterations of peroxisome proliferator-activated receptor δ activity affect fatty acid-controlled adipose differentiation. J. Biol. Chem. 2000;275:38768–38773. doi: 10.1074/jbc.M006450200. [DOI] [PubMed] [Google Scholar]
- 152.Matsusue K., Peters J. M., Gonzalez F. J. PPARβ/δ potentiates PPARγ-stimulated adipocyte differentiation. FASEB J. 2004;18:1477–1479. doi: 10.1096/fj.04-1944fje. [DOI] [PubMed] [Google Scholar]
- 153.Wang Y. X., Lee C. H., Tiep S., Yu R. T., Ham J., Kang H., Evans R. M. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 154.Barak Y., Liao D., He W., Ong E. S., Nelson M. C., Olefsky J. M., Boland R., Evans R. M. Effects of peroxisome proliferator-activated receptor δ on placentation, adiposity, and colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 2002;99:303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li A. C., Glass C. K. PPAR- and LXR-dependent pathways controlling lipid metabolism and the development of atherosclerosis. J. Lipid Res. 2004;45:2161–2173. doi: 10.1194/jlr.R400010-JLR200. [DOI] [PubMed] [Google Scholar]
- 156.Ross S. E., Erickson R. L., Gerin I., DeRose P. M., Bajnok L., Longo K. A., Misek D. E., Kuick R., Hanash S. M., Atkins K. B., et al. Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor α in adipocyte metabolism. Mol. Cell. Biol. 2002;22:5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Juvet L. K., Andresen S. M., Schuster G. U., Dalen K. T., Tobin K. A., Hollung K., Haugen F., Jacinto S., Ulven S. M., Bamberg K., et al. On the role of liver X receptors in lipid accumulation in adipocytes. Mol. Endocrinol. 2003;17:172–182. doi: 10.1210/me.2001-0210. [DOI] [PubMed] [Google Scholar]
- 158.Peet D. J., Turley S. D., Ma W. Z., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 159.Hummasti S., Laffitte B. A., Watson M. A., Galardi C., Chao L. C., Ramamurthy L., Moore J. T., Tontonoz P. Liver X receptors are regulators of adipocyte gene expression but not differentiation: identification of apoD as a direct target. J. Lipid Res. 2004;45:616–625. doi: 10.1194/jlr.M300312-JLR200. [DOI] [PubMed] [Google Scholar]
- 160.Seo J. B., Moon H. M., Kim W. S., Lee Y. S., Jeong H. W., Yoo E. J., Ham J., Kang H., Park M. G., Steffensen K. R., et al. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor γ expression. Mol. Cell. Biol. 2004;24:3430–3444. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Laffitte B. A., Repa J. J., Joseph S. B., Wilpitz D. C., Kast H. R., Mangelsdorf D. J., Tontonoz P. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. U.S.A. 2001;98:507–512. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Laffitte B. A., Chao L. C., Li J., Walczak R., Hummasti S., Joseph S. B., Castrillo A., Wilpitz D. C., Mangelsdorf D. J., Collins J. L., et al. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5419–5424. doi: 10.1073/pnas.0830671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dalen K. T., Ulven S. M., Bamberg K., Gustafsson J. A., Nebb H. I. Expression of the insulin-responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor α. J. Biol. Chem. 2003;278:48283–48291. doi: 10.1074/jbc.M302287200. [DOI] [PubMed] [Google Scholar]
- 164.Ulven S. M., Dalen K. T., Gustafsson J. A., Nebb H. I. Tissue-specific autoregulation of the LXRα gene facilitates induction of apoE in mouse adipose tissue. J. Lipid Res. 2004;45:2052–2062. doi: 10.1194/jlr.M400119-JLR200. [DOI] [PubMed] [Google Scholar]
- 165.Gerin I., Dolinsky V. W., Shackman J. G., Kennedy R. T., Chiang S. H., Burant C. F., Steffensen K. R., Gustafsson J. A., MacDougald O. A. LXRβ is required for adipocyte growth, glucose homeostasis, and β-cell function. J. Biol. Chem. 2005;280:23024–23031. doi: 10.1074/jbc.M412564200. [DOI] [PubMed] [Google Scholar]
- 166.Kalaany N. Y., Gauthier K. C., Zavacki A. M., Mammen P. P., Kitazume T., Peterson J. A., Horton J. D., Garry D. J., Bianco A. C., Mangelsdorf D. J. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 2005;1:231–244. doi: 10.1016/j.cmet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 167.Stulnig T. M., Steffensen K. R., Gao H., Reimers M., Dahlman-Wright K., Schuster G. U., Gustafsson J. A. Novel roles of liver X receptors exposed by gene expression profiling in liver and adipose tissue. Mol. Pharmacol. 2002;62:1299–1305. doi: 10.1124/mol.62.6.1299. [DOI] [PubMed] [Google Scholar]
- 168.Bavner A., Sanyal S., Gustafsson J. A., Treuter E. Transcriptional corepression by SHP: molecular mechanisms and physiological consequences. Trends Endocrinol. Metab. 2005;16:478–488. doi: 10.1016/j.tem.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 169.Wang L., Liu J., Saha P., Huang J., Chan L., Spiegelman B., Moore D. D. The orphan nuclear receptor SHP regulates PGC-1α expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 170.Darlington G. J., Ross S. E., MacDougald O. A. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 171.Carmona M. C., Hondares E., Rodriguez de la Concepcion M. L., Rodriguez-Sureda V., Peinado-Onsurbe J., Poli V., Iglesias R., Villarroya F., Giralt M. Defective thermoregulation, impaired lipid metabolism, but preserved adrenergic induction of gene expression in brown fat of mice lacking C/EBPβ. Biochem. J. 2005;389:47–56. doi: 10.1042/BJ20050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tanaka T., Yoshida N., Kishimoto T., Akira S. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang N. D., Finegold M. J., Bradley A., Ou C. N., Abdelsayed S. V., Wilde M. D., Taylor L. R., Wilson D. R., Darlington G. J. Impaired energy homeostasis in C/EBPα knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 174.Flodby P., Barlow C., Kylefjord H., Ahrlund-Richter L., Xanthopoulos K. G. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein α. J. Biol. Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 175.Carmona M. C., Iglesias R., Obregon M. J., Darlington G. J., Villarroya F., Giralt M. Mitochondrial biogenesis and thyroid status maturation in brown fat require CCAAT/enhancer-binding protein α. J. Biol. Chem. 2002;277:21489–21498. doi: 10.1074/jbc.M201710200. [DOI] [PubMed] [Google Scholar]
- 176.Linhart H. G., Ishimura-Oka K., DeMayo F., Kibe T., Repka D., Poindexter B., Bick R. J., Darlington G. J. C/EBPα is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12532–12537. doi: 10.1073/pnas.211416898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yang J., Croniger C. M., Lekstrom-Himes J., Zhang P., Fenyus M., Tenen D. G., Darlington G. J., Hanson R. W. Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein α. J. Biol. Chem. 2005;280:38689–38699. doi: 10.1074/jbc.M503486200. [DOI] [PubMed] [Google Scholar]
- 178.Porse B. T., Pedersen T. A., Xu X., Lindberg B., Wewer U. M., Friis-Hansen L., Nerlov C. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–258. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- 179.Chen S. S., Chen J. F., Johnson P. F., Muppala V., Lee Y. H. C/EBPβ, when expressed from the C/EBPα gene locus, can functionally replace C/EBPα in liver but not in adipose tissue. Mol. Cell. Biol. 2000;20:7292–7299. doi: 10.1128/mcb.20.19.7292-7299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.White M. F. IRS proteins and the common path to diabetes. Am. J. Physiol. Endocrinol. Metab. 2002;283:E413–E422. doi: 10.1152/ajpendo.00514.2001. [DOI] [PubMed] [Google Scholar]
- 181.Laustsen P. G., Michael M. D., Crute B. E., Cohen S. E., Ueki K., Kulkarni R. N., Keller S. R., Lienhard G. E., Kahn C. R. Lipoatrophic diabetes in Irs1−/−/Irs3−/− double knockout mice. Genes Dev. 2002;16:3213–3222. doi: 10.1101/gad.1034802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tseng Y. H., Kriauciunas K. M., Kokkotou E., Kahn C. R. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol. Cell. Biol. 2004;24:1918–1929. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tseng Y. H., Butte A. J., Kokkotou E., Yechoor V. K., Taniguchi C. M., Kriauciunas K. M., Cypess A. M., Niinobe M., Yoshikawa K., Patti M. E., Kahn C. R. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat. Cell Biol. 2005;7:601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- 184.Shima H., Pende M., Chen Y., Fumagalli S., Thomas G., Kozma S. C. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Volarevic S., Stewart M. J., Ledermann B., Zilberman F., Terracciano L., Montini E., Grompe M., Kozma S. C., Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40 S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 186.Fingar D. C., Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 187.Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L., Mestan J., Mueller M., Fumagalli S., Kozma S. C., Thomas G. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pende M., Kozma S. C., Jaquet M., Oorschot V., Burcelin R., Le Marchand-Brustel Y., Klumperman J., Thorens B., Thomas G. Hypoinsulinaemia, glucose intolerance and diminished β-cell size in S6K1-deficient mice. Nature (London) 2000;408:994–997. doi: 10.1038/35050135. [DOI] [PubMed] [Google Scholar]
- 189.Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature (London) 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 190.Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., et al. The TSC1–2 tumor suppressor controls insulin-P13K signaling via regulation of IRS proteins. J. Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lorenzo M., Valverde A. M., Teruel T., Benito M. IGF-I is a mitogen involved in differentiation-related gene expression in fetal rat brown adipocytes. J. Cell Biol. 1993;123:1567–1575. doi: 10.1083/jcb.123.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Valverde A. M., Kahn C. R., Benito M. Insulin signaling in insulin receptor substrate (IRS)-1-deficient brown adipocytes requirement of IRS-1 for lipid synthesis. Diabetes. 1999;48:2122–2131. doi: 10.2337/diabetes.48.11.2122. [DOI] [PubMed] [Google Scholar]
- 193.Fasshauer M., Klein J., Kriauciunas K. M., Ueki K., Benito M., Kahn C. R. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol. Cell. Biol. 2001;21:319–329. doi: 10.1128/MCB.21.1.319-329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mur C., Arribas M., Benito M., Valverde A. M. Essential role of insulin-like growth factor I receptor in insulin-induced fetal brown adipocyte differentiation. Endocrinology. 2003;144:581–593. doi: 10.1210/en.2002-220828. [DOI] [PubMed] [Google Scholar]
- 195.Valverde A. M., Lorenzo M., Navarro P., Benito M. Phosphatidylinositol 3-kinase is a requirement for insulin-like growth factor I-induced differentiation, but not for mitogenesis, in fetal brown adipocytes. Mol. Endocrinol. 1997;11:595–607. doi: 10.1210/mend.11.5.9924. [DOI] [PubMed] [Google Scholar]
- 196.Valverde A. M., Benito M., Lorenzo M. The brown adipose cell: a model for understanding the molecular mechanisms of insulin resistance. Acta Physiol. Scand. 2005;183:59–73. doi: 10.1111/j.1365-201X.2004.01384.x. [DOI] [PubMed] [Google Scholar]
- 197.Yeh W. C., Bierer B. E., Mcknight S. L. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11086–11090. doi: 10.1073/pnas.92.24.11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tsukiyama-Kohara K., Poulin F., Kohara M., DeMaria C. T., Cheng A., Wu Z., Gingras A. C., Katsume A., Elchebly M., Spiegelman B. M., et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- 199.Lehr L., Kuehne F., Arboit P., Giacobino J. P., Poulin F., Muzzin P., Jimenez M. Control of 4E-BP1 expression in mouse brown adipose tissue by the β3-adrenoceptor. FEBS Lett. 2004;576:179–182. doi: 10.1016/j.febslet.2004.08.081. [DOI] [PubMed] [Google Scholar]
- 200.Lipinski M. M., Jacks T. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- 201.Hansen J. B., te Riele H., Kristiansen K. Novel function of the retinoblastoma protein in fat: regulation of white versus brown adipocyte differentiation. Cell Cycle. 2004;3:774–778. [PubMed] [Google Scholar]
- 202.Hansen J. B., Kristiansen K. Pocket proteins control white versus brown fat cell differentiation. Cell Cycle. 2006;5:341–342. doi: 10.4161/cc.5.4.2465. [DOI] [PubMed] [Google Scholar]
- 203.Cherington V., Morgan B., Spiegelman B. M., Roberts T. M. Recombinant retroviruses that transduce individual polyoma tumor antigens: effects on growth and differentiation. Proc. Natl. Acad. Sci. U.S.A. 1986;83:4307–4311. doi: 10.1073/pnas.83.12.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cherington V., Brown M., Paucha E., St Louis J., Spiegelman B. M., Roberts T. M. Separation of simian virus 40 large-T-antigen-transforming and origin-binding functions from the ability to block differentiation. Mol. Cell. Biol. 1988;8:1380–1384. doi: 10.1128/mcb.8.3.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ali S. H., DeCaprio J. A. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 2001;11:15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- 206.Scimè A., Grenier G., Huh M. S., Gillespie M. A., Bevilacqua L., Harper M. E., Rudnicki M. A. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1α. Cell Metab. 2005;2:283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 207.Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L., MacDougald O. A. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 208.Longo K. A., Wright W. S., Kang S., Gerin I., Chiang S. H., Lucas P. C., Opp M. R., MacDougald O. A. Wnt10b inhibits development of white and brown adipose tissues. J. Biol. Chem. 2004;279:35503–35509. doi: 10.1074/jbc.M402937200. [DOI] [PubMed] [Google Scholar]
- 209.Rodriguez de la Concepcion M. L., Yubero P., Iglesias R., Giralt M., Villarroya F. Lithium inhibits brown adipocyte differentiation. FEBS Lett. 2005;579:1670–1674. doi: 10.1016/j.febslet.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 210.Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 211.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado de Oliveira R., Leid M., McBurney M. W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPARγ. Nature (London) 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Moncada S., Erusalimsky J. D. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat. Rev. Mol. Cell Biol. 2002;3:214–220. doi: 10.1038/nrm762. [DOI] [PubMed] [Google Scholar]
- 213.Nisoli E., Clementi E., Tonello C., Sciorati C., Briscini L., Carruba M. O. Effects of nitric oxide on proliferation and differentiation of rat brown adipocytes in primary cultures. Br. J. Pharmacol. 1998;125:888–894. doi: 10.1038/sj.bjp.0702131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nisoli E., Clementi E., Paolucci C., Cozzi V., Tonello C., Sciorati C., Bracale R., Valerio A., Francolini M., Moncada S., Carruba M. O. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 215.Oishi Y., Manabe I., Tobe K., Tsushima K., Shindo T., Fujiu K., Nishimura G., Maemura K., Yamauchi T., Kubota N. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 216.Chen Z., Torrens J. I., Anand A., Spiegelman B. M., Friedman J. M. Krox20 stimulates adipogenesis via C/EBPβ-dependent and -independent mechanisms. Cell Metab. 2005;1:93–106. doi: 10.1016/j.cmet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 217.Tong Q., Dalgin G., Xu H., Ting C. N., Leiden J. M., Hotamisligil G. S. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 218.Tsai J., Tong Q., Tan G., Chang A. N., Orkin S. H., Hotamisligil G. S. The transcription factor GATA2 regulates differentiation of brown adipocytes. EMBO Rep. 2005;6:879–884. doi: 10.1038/sj.embor.7400490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ge K., Guermah M., Yuan C. X., Ito M., Wallberg A. E., Spiegelman B. M., Roeder R. G. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature (London) 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 220.Castillo G., Brun R. P., Rosenfield J. K., Hauser S., Park C. W., Troy A. E., Wright M. E., Spiegelman B. M. An adipogenic cofactor bound by the differentiation domain of PPARγ. EMBO J. 1999;18:3676–3687. doi: 10.1093/emboj/18.13.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]