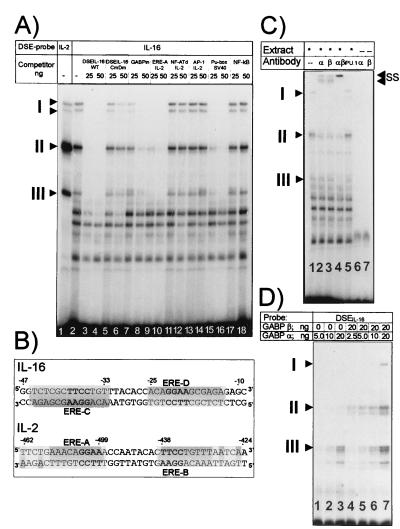
Figure 4.
GABP factors bind to the Ets-like sequence motifs of IL-16 promoter. (A) EMSAs using nuclear proteins from Jurkat cells and the DSE from the IL-16 promoter as probe. Two micrograms of nuclear protein from noninduced Jurkat cells was incubated either with the DSEIL-2 (lane 1) or DSEIL-16 (lanes 2–18) as probes (see B) followed by electrophoresis on a native 4% polyacrylamide gel. For competition, 25 and 50 ng of the following oligonucleotides were added: lanes 3 and 4, DSEIL-16; lanes 5 and 6, DSEIL-16 mutated in both Ets motifs; lanes 7 and 8, IL-16 Ets motif at position +12; lanes 9 and 10, IL-2 ERE-A motif (see B); lanes 11 and 12, distal NF-AT site from the murine IL-2 promoter (see ref. 22); lanes 13 and 14, AP-1 binding site from the IL-2 promoter (22); lanes 15 and 16, Ets site from the SV40 enhancer (20); lanes 17 and 18, consensus NF-κB site. (B) Sequences of DSEs from the IL-16 promoter and distal IL-2 enhancer (20). The palindromic organization of Ets-related elements, EREs, is indicated in gray. (C) Supershift EMSAs with Abs raised against GABPα and -β. In lanes 1–5, 1 μg of nuclear protein from Jurkat cells was incubated with the DSEIL-16 probe, alone or with 1 μg of Ab raised against GABPα (lane 2), -β (lane 3), or with both Abs (lane 4). As a control, a Pu.1-specific Ab was added in lane 5. In lanes 6 and 7, the DSEIL-16 probe was incubated with Abs alone. SS, complexes supershifted by GABP Abs. (D) Binding of recombinant GABPα and -β to the DSEIL-16. Bacterially expressed GABPα or -β (2.5–20 ng) was incubated with a DSEIL-16 probe as indicated. I–III indicate DNA–protein complexes identical in mobility to those generated with nuclear proteins from Jurkat cells (A).