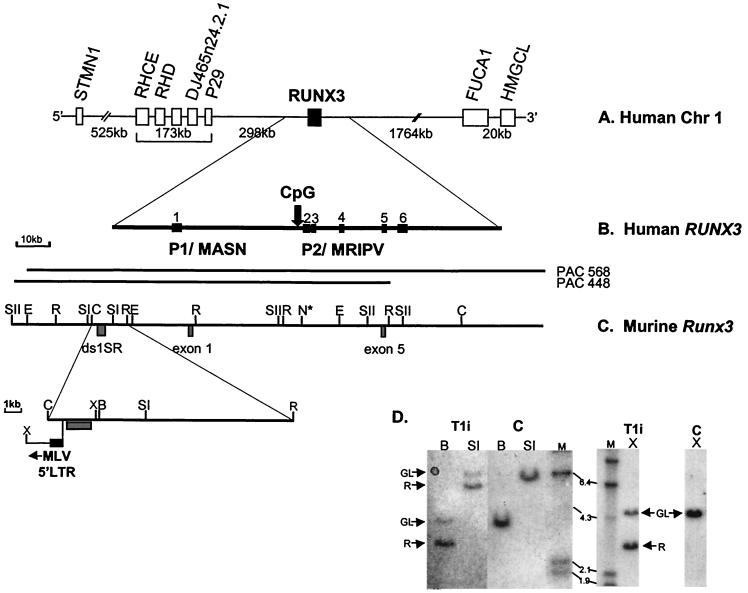
FIG. 2.
Comparative physical map of human RUNX3 and murine Runx3 genes. (A) Physical map of human chromosome 1 spanning 2.7 Mb around the RUNX3 gene, based on public database information (http://genome.ucsc.edu). All of the genes indicated are transcribed in the same orientation as RUNX3. (B) Genomic structure of the human RUNX3 gene. The locations of the distal P1 and proximal P2 promoter regions are indicated. Vertical arrow, P2-associated CpG island. (C) Physical map of murine Runx3 gene. PAC 448-L9 and PAC568-D12 were isolated from the murine RPC121 PAC library. ds1-SR and the Runx3 exon 1 and exon 5 probes are indicated. Only restriction sites mapped with the probes are shown. N* indicates the presence of multiple NotI sites and the inferred position of P2-associated CpG island from draft murine genome sequence (accession number AF169246). Expanded region shows the position of the integrated MLV in T1i. Restriction enzyme abbreviations: SI, SstI; SII, SstII; E, EagI; N, NotI; C, ClaI; R, EcoRI; X, XhoI; B, BamHI. (D) Southern analysis of MLV-infected CD2-MYC lymphomas. Genomic DNA (20 μg; T1i cell line and control [C] nontransgenic thymus) was digested with BamHI (B), SstI (SI), and XhoI (X) and hybridized with the ds1-SR probe. GL, germ line; R, rearranging fragment. HindIII-digested λ DNA was used as molecular size markers (sizes shown in kilobases).