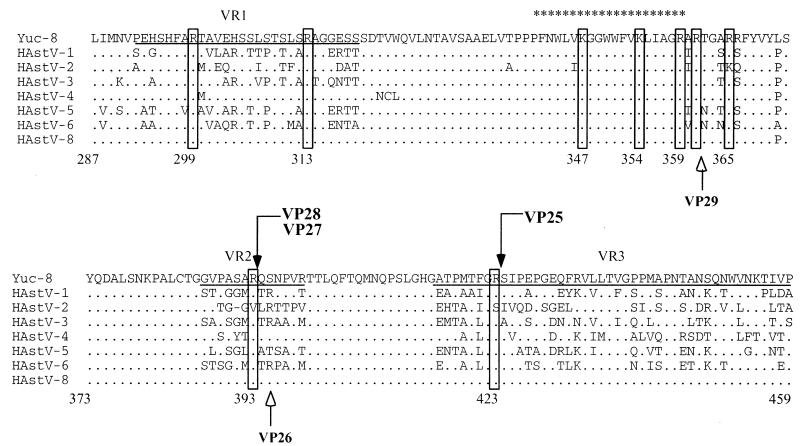
FIG. 4.
N-terminal sequence of VP28-derived proteins. Purified Yuc8 particles were trypsin treated and electroblotted to PVDF membranes. VP28, VP27, and VP25 were purified, and the N termini were sequenced. The sequence alignment of eight HAstV serotypes between residues 287 and 459 of ORF2 is shown. The cleavage sites for the proteins VP28, VP27, and VP25 (boxes marked by black arrows) and the amino acid residues which could potentially represent the carboxy end of the VP41-derived proteins (boxes at the amino acid residues 299, 313, 347, 354, 359, 361, and 365) are indicated. The cleavage sites for the proteins VP29 and VP26 of HAstV-2, previously described (20), are also indicated (white arrows). The dots along the sequence denote identity, and only amino acid changes are marked. The previously described variable regions VR1 and VR2 and part of VR3 (16) are underlined. The common astrovirus epitope predicted (21) is marked with asterisks. The numbers below the sequences indicate the amino acid position based on the Yuc8 sequence. Sequence alignment was made by Clustalw analysis (http://www.ebi.ac.uk/clustalw/) using sequences with accession numbers L23513 (HAstV-1), A45695 (HAstV-2), AF141381(HAstV-3), Z33883 (HAstV-4), U15136 (HAstV-5), Z46658 (HAstV-6), Z66541 (HAstV-8), and AF260508 (Yuc8).