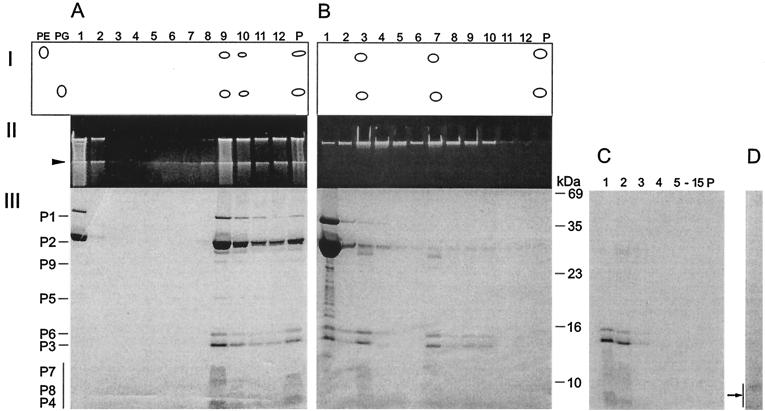
FIG. 5.
Dissociation of PM2 particles with urea and separation of the products by rate zonal centrifugation (see Materials and Methods). (A) Untreated particles. (B) Particles dissociated with 4.5 M urea. (Panel I) Major lipids separated by thin-layer chromatography with PE and PG as standards. (Panel II) Viral DNA in upper SDS-polyacylamide gel stained with ethidium bromide. (Panel III) Structural proteins in Coomassie brilliant blue-stained SDS-polyacrylamide gel. P, pellet. The boundary between the upper gel and the lower gel is indicated by an arrowhead. The positions of the PM2 structural proteins are indicated on the left (the vertical bar denotes the small membrane proteins forming diffuse bands). The numbers to the right of panel B indicate the molecular masses of the standard proteins. (C) Isolated phage lipid cores in 4.5 M urea were solubilized with 0.1% SDS and analyzed with a rate zonal sucrose gradient containing 2 M urea, 0.1% SDS, and PM2 buffer. Fifteen fractions and the pellet were collected and analyzed by SDS-PAGE. The area indicated by an arrow was subjected to N-terminal amino acid sequencing. (D) Isolated phage lipid cores in 4.5 M urea were treated with trypsin (50 μg/ml) and soybean trypsin inhibitor (500 μg/ml) and analyzed with a rate zonal sucrose gradient (PM2 buffer). One visible light-scattering zone was detected and analyzed by SDS-PAGE. No other PM2 proteins were detected in the gradient fractions.