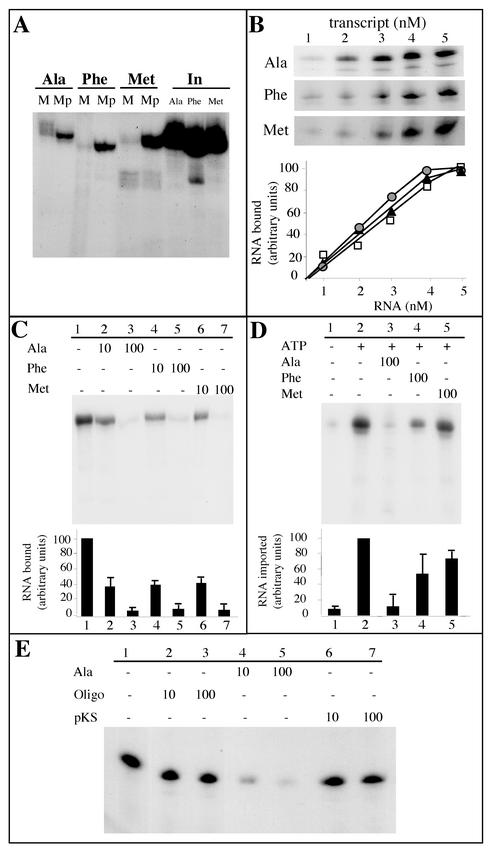
FIG. 9.
Analysis of tRNAAla, tRNAPhe, and tRNAMet-e binding to mitochondria or import into mitochondria. (A) Labeled in vitro-transcribed tRNAAla, tRNAPhe, and tRNAMet (2 × 105 cpm each) were incubated under standard import conditions with intact mitochondria (M) or with mitoplasts (Mp). In, input RNAs (5,000 cpm). (B) Bound tRNAAla, tRNAPhe, and tRNAMet-e as a function of tRNAAla, tRNAPhe, and tRNAMet-e concentration, respectively. Increasing concentrations of labeled tRNA transcripts (from 1 to 5 nM) were incubated with isolated mitochondria (corresponding to 100 μg of proteins). Incubation was performed for 10 min under the same conditions as for tRNA import, except that the RNase digestion step was omitted. (C) Competition experiments of tRNAAla binding onto isolated mitochondria using unlabeled tRNAAla, tRNAPhe, or tRNAMet-e as competitors. For these experiments, isolated mitochondria (corresponding to 100 μg of proteins) were preincubated, for 10 min on ice, in the absence of competitor (−) or in the presence of a 10-fold or a 100-fold excess of competitor. Labeled tRNAAla (105 cpm) was then added, and incubation was prolonged for 10 min under the same conditions as for tRNA import, except that the RNase digestion step was omitted. (D) Competition experiments of mitochondrial tRNAAla import into isolated mitochondria using unlabeled tRNAAla, tRNAPhe, or tRNAMet-e as competitor. For these experiments, isolated mitochondria (corresponding to 400 μg of proteins) were preincubated, for 10 min on ice, in the absence of competitor (−) or in the presence of a 100-fold excess of competitor. Then, 2 × 105 cpm of labeled tRNAAla was added, and incubation was prolonged for 15 min under the same conditions as for tRNA import. The experiments were performed in the absence (−) or in the presence (+) of 5 mM ATP. After incubation, a proteinase K treatment was performed as described in Materials and Methods prior to digestion with RNases. (E) Competition experiments of tRNAAla binding to isolated mitochondria using an unlabeled oligodeoxyribonucleotide (Oligo) corresponding to the complete sequence of the cytosol-specific tRNAMet-e (Fig. 8A) or AluI-digested pBluescript vector (pKS) as competitors. The competition experiments were conducted essentially as described for panel C. For graphical representation of the results in panels B, C, and D, the amounts of bound or protected RNAs were quantified using a phosphorimager. Bars represent standard deviations from two and three independent experiments for panels C and D, respectively, and mean values were used to plot the graph.