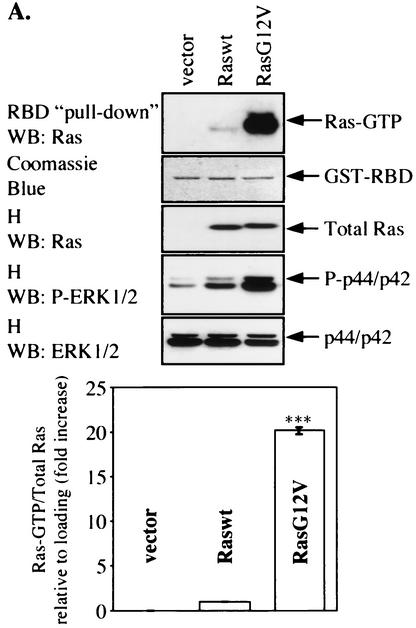
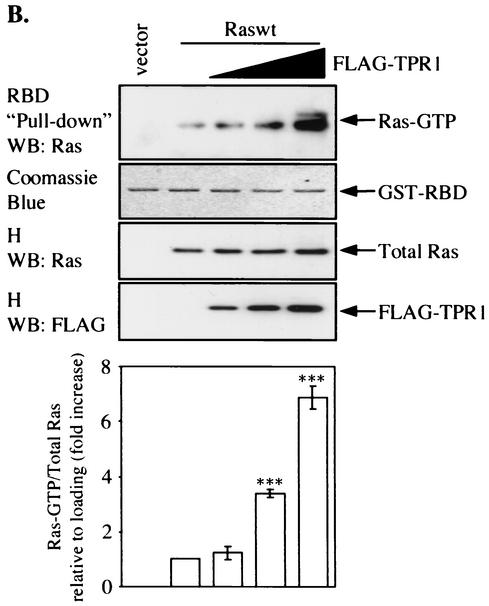
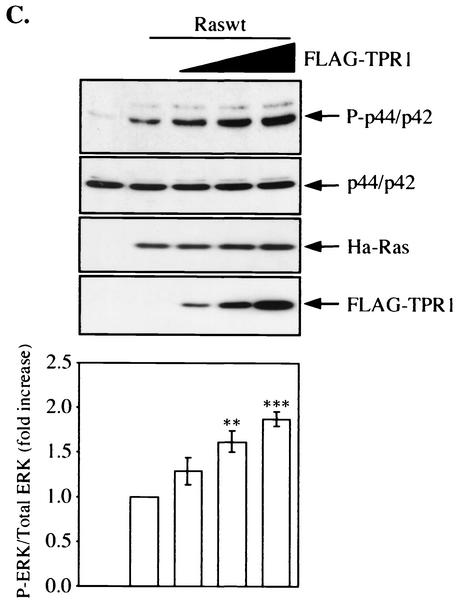
FIG. 6.
Involvement of TPR1 in accumulation of active Ras and phosphorylation of ERKs. (A) HeLa cells were transiently transfected with empty vector or with the Raswt or RasG12V expression construct. The cells were serum starved for 24 h, and Ras activity was measured using the RBD of Raf-1. The cell lysates were precipitated with the GST-RBD fusion protein coupled to glutathione-Sepharose beads. The bound Ras-GTP was detected in the precipitates by Western blotting (WB) using an anti-Ras Ab (top panel). The GST-RBD fusion protein was detected by Coomassie blue staining (second panel from top) and serves as a loading control. The total amount of Ras protein in each sample before GST-RBD binding was determined by probing the cell homogenates (H) with an anti-Ras Ab (middle panel). The homogenates were also assayed for any changes in the phosphorylation of the intrinsic ERK1/2 (P-ERK1/2) (second panel from bottom) and compared to the nonphosphorylated form of the two kinase isoforms (bottom panel). (B) Raswt was expressed alone (second lane from the left) or together with increasing amounts of FLAG-TPR1 in HeLa cells. As in panel A, after a 24-h serum starvation period, active Ras (top panel) was precipitated from the cell homogenates (H) using the GST-RBD affinity reagent. (C) The TPR1-dependent ERK1/2 phosphorylation was analyzed in cells transfected under the same conditions. Results shown are representative of at least three independent experiments. Quantitative analyses of all blots were performed using the ImageQuant program (Molecular Dynamics). For each individual experiment, values for the relative amounts of Ras-GTP were calculated as fold increase compared to the basal level Raswt condition, which was assigned a value of 1. The bars in the histograms are the means ± standard errors of the means for three experiments (n = 3). Values that were significantly different from the basal level are indicated (**, P < 0.01; ***, P < 0.001).