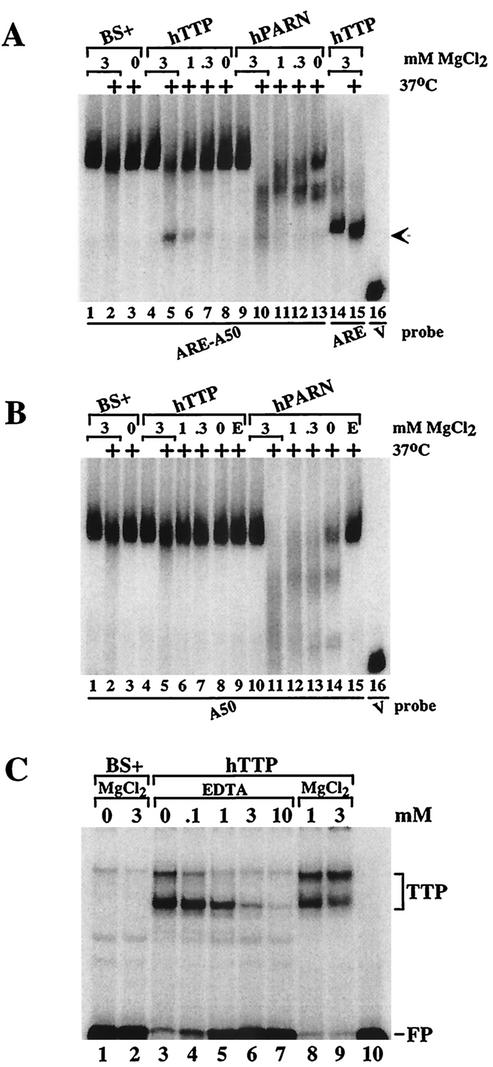
FIG. 5.
Characterization of the TTP-induced deadenylating activity. Extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), and CMV.hPARN.flag (hPARN) were incubated on ice or at 37°C (+) for 60 min and processed as described for Fig. 2. (A) MgCl2 was included in the reaction mixture to final concentrations of 3 mM (lanes 1 and 2, lanes 4 and 5, lanes 9 and 10, and lanes 14 and 15), or 1 mM (lanes 6 and 11), 0.3 mM (lanes 7 and 12) or was not included (lanes 3, 8, and 13), and the mixtures were incubated and processed as described for Fig. 2. The arrow indicates the migration positions of the deadenylated product of probe ARE-A50 (lanes 5 to 7) and the ARE probe (lanes 14 and 15). The position of probe V migration is shown in lane 16. (B) MgCl2 was included in the reaction mixture to final concentrations of 3 mM (lanes 1 and 2, lanes 4 and 5, and lanes 10 and 11), 1 mM (lanes 6 and 12), or 0.3 mM (lanes 7 and 13) or was not included (lanes 3, 8, and 13), and the mixtures were incubated and processed as described for Fig. 2. EDTA (1 mM) was present during the incubation in lanes 9 and 15. The position of probe V migration is shown in lane 16. (C) Extracts from 293 cells transfected with vector alone (BS+) or CMV.hTTP.tag (hTTP) were incubated with probe ARE-A50 in the absence (lanes 1 and 3) or presence of MgCl2 (3 mM, lanes 2 and 9; 1 mM, lane 8) or with increasing concentrations of EDTA (0 to 10 mM, lanes 3 to 7); the reactions were then used in a gel shift assay. Lane 10 (P′) was loaded with probe alone (RNase T1 digested). The migration positions of the TTP-RNA complexes (TTP) and the free probe (FP) are indicated.