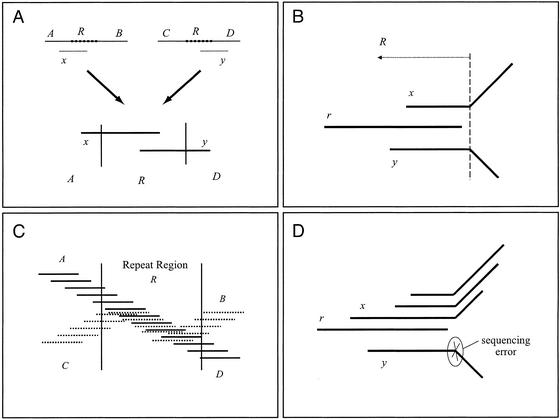
Figure 3.
Contig assembly. (A) How merging reads across the boundary of a repeat may result in a misassembly. Regions A, B, C, and D are unique regions, and region R is a repeat occurring twice in the genome. Reads x and y overlap in region R. Thus, regions A and D are wrongly joined after merging reads x and y. (B) A potential repeat boundary. Read r overlaps both reads x and y, but reads x and y do not overlap each other; they disagree in their rightmost ends. Here, a repeat R starting inside reads x and y and including the full read r is shown. In practice, sequencing errors rather than repeats often cause such patterns of overlap. (C) Contigs are created by merging reads up to the potential boundaries of repeats. A potential repeat boundary is any place where a read may be extended with two nonoverlapping reads. Two regions of the genome covered with reads are shown here. One region (A-R-D) is covered with solid line reads and the second region (C-R-B) with dotted line reads. The two regions meet in the repeat R creating five contigs: these are the unique contigs corresponding to unique sequences A, B, C, and D, and the repeat contig corresponding to the repeat R, where reads from both copies of R are overcollapsed into one contig. According to the algorithm used to construct contigs, the contig corresponding to R would have exactly the reads that are fully included in the boundaries of R. All the other reads would be assigned to contigs A, B, C, and D. (D) Sequencing errors. Read r dominates read y because the neighbors of y are all neighbors of r. This is caused by a sequencing error on y, which is marked in the figure. Note that if y represented correct sequence, it would likely be extended to the right by some read that did not overlap r, and thus r would not dominate y.