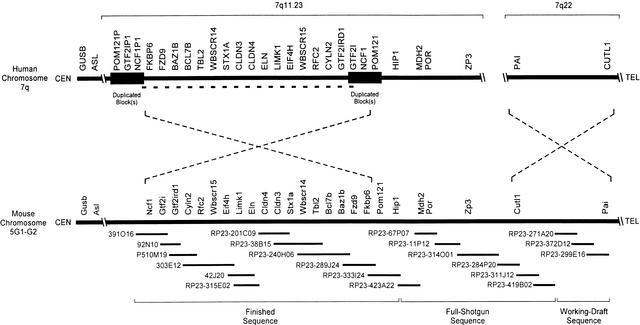
Figure 1.
Long-range organization of human and mouse Williams syndrome (WS) regions. A physical map of the WS regions on human chromosome 7q and mouse chromosome 5G is depicted emphasizing the positions of the known genes residing within and flanking the interval commonly deleted in WS (DeSilva et al. 1999; Francke 1999; Hockenhull et al. 1999; Osborne 1999; Korenberg et al. 2000; Peoples et al. 2000; Valero et al. 2000). In the human WS region, this interval spans ∼1.6 Mb (indicated by a bold dashed line) and is flanked by duplicated blocks of DNA of near-identical sequence (estimated at ∼300 kb in size; indicated by dark rectangles). The relative positions of the centromere (CEN) and telomere (TEL) are indicated in each case. Note the inverted orientation of the two discontiguous segments of human chromosome 7 relative to the single contiguous segment of mouse chromosome 5G. The relative positions of the known human and mouse genes residing in this region are indicated, with additional details provided in Table 1. Depicted below the map of the mouse WS region are the 21 overlapping BAC/PAC clones selected for sequencing (see http://bio.cse.psu.edu/publications/desilva for a complete contig map of the mouse WS region), with the current sequencing status (finished, full shotgun, or working draft) indicated at the bottom (also see Table 2). Note that the depicted genomic regions and the BAC/PAC clones are not drawn to scale.