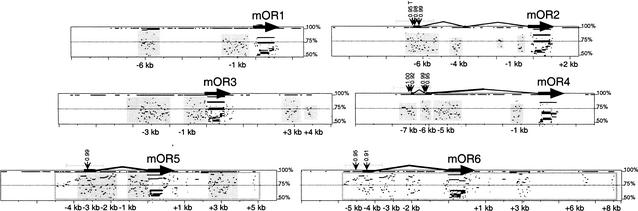
Figure 4.
PIPmaker analysis illustrating genomic sequence similarity in the vicinity of the mouse OR genes. Gene homology in the vicinity of the six mouse OR genes is plotted by PipMaker (Schwartz et al. 2000). Coding sequences are shown as thick black arrows. Upstream exons (horizontal) and introns (bent) as determined by 5′ RACE-PCR are shown as thin black lines. Positions relative to translation start are shown in kilobases (kb). Gene structures for mOR5 (= mOR10), mOR6 (= mOR28), and mOR4 (= mOR83) isoforms are identical to those reported in Tsuboi et al. (1999). Each gene was compared to all other mouse and human OR genes at the TCR-α/δ loci. Detected sequence homology (>50% identity) is plotted according to position in the test gene and level of sequence similarity. Light grey boxes surround homology detected exclusively between a mouse OR gene and its human ortholog. The only paralogous homology detected is between mOR5 and mOR6 (because of a block duplication), and this block of homology for both genes is surrounded by grey-framed, open rectangles. Regions ± 1 kb from transcription start sites (grey horizontal bars) were analyzed for promoter signals. Positions of NNPP promoter hits with scores >0.90 are shown (suffix “T” indicates TATA, no suffix indicates non-TATA). Positions of RepeatMasker sequences are indicated as breaks in horizontal black lines at the top (100% identity level) of each box.