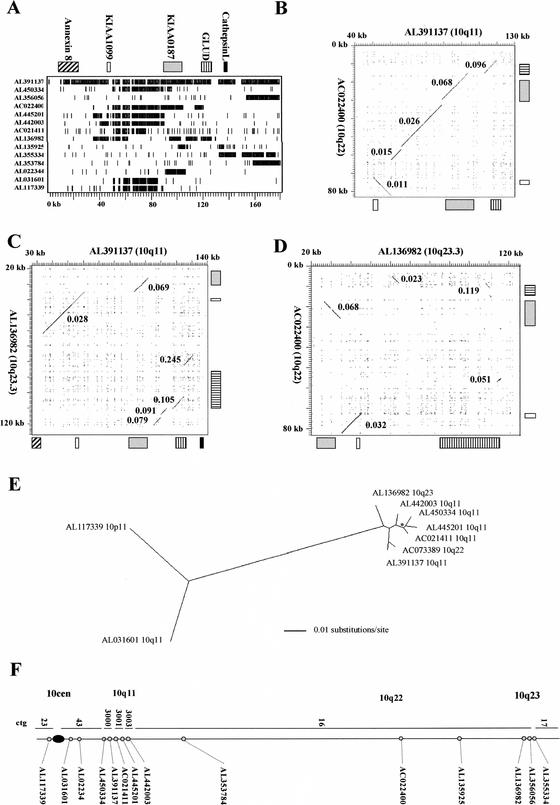
Figure 4.
Analysis of pseudogene clusters containing KIAA0187 paralogs. (A) Human genomic blast hits to AL391137. The position of all independent human clones with >90% sequence identity to AL391137 over >2 kb are shown relative to RepeatMasked AL391137 sequence (hit 1 is the self-comparison). The positions of gene-related sequences within AL391137 are indicated above, with the scale in kilobases indicated below. Numbers refer to the clones shown in F. The GLUD1-related sequences identified include the functional GLUD1 gene (within hit 8) and two nonprocessed pseudogene fragments, one of which (GLUDP3 within hit 4) has been previously mapped to 10q22 (Deloukas et al. 1993). The Cathepsin L–related sequences include two pseudogenes, CTSLL1 and CTSLL1-2 (within hits 10 and 1, respectively), which have been mapped previously to 10q (Bryce et al. 1994). (B–D) Dot matrix analyses of 10q pseudogene clusters. The positions of gene-related sequences are shown for each clone. Kimura 2 parameter distances for each individual region of high identity are indicated. The highly diverged match in C (K2P = 0.245) is owing to a cluster of Alu elements and is assumed to be coincidental. (E) Maximum-likelihood tree of KIAA1099 paralogs generated from a 10952-bp alignment spanning nucleotides 55178–59761 of AL391137. The scale (in substitutions/site) for the branch lengths is shown. All branchpoints have >95% bootstrap support with a single exception (asterisk), which has 81% support. (F) Distribution of sequences related to AL391137 on chromosome 10. The position of each clone identified in the BLAST analysis within Sanger Centre contigs (Bentley et al. 2001) is shown. Cytogenetic locations (established by FISH; Table 1) are also shown.