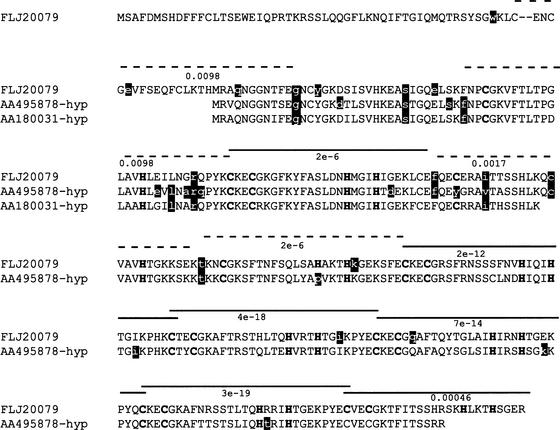
Figure 1.
Damaging mutations in redundant motifs. FLJ20079 is aligned with inferred proteins AA495878-hyp and AA180031-hyp. Protein sequences were inferred by obtaining the EST sequences AA495878 and AA180031 from which the dbSNP variants were derived. AA180031 and AA495878 were only 93% and 91% identical to the gene encoding FLJ20079, respectively, but 100% identical to other regions of the human genome. Genomic sequences surrounding the region that matched AA180031 and AA495878 were retrieved and translated. The protein sequences were aligned to FLJ20079 and the start of the proteins interpreted to be the first Met that aligned to FLJ20079. The inferred hypothetical sequences were named AA180031-hyp and AA495878-hyp, from the ESTs from which they were derived. The nonsynonymous/synonymous ratio (Yang 1997) for AA180031-hyp and AA495878-hyp with FLJ20079 is 0.50 and 0.55, respectively, indicating that these proteins are undergoing purifying selection and may be functional. The three proteins contain zinc-finger motifs. Each putative zinc finger is indicated by a dashed or solid line and the value beneath the line is the e-value score for the region from FLJ20079 with the C2H2 zinc-finger motif (IPB000822) (Henikoff and Henikoff 1994). Regions with solid lines on top have the Cys and His residues involved in binding the zinc atom conserved in the three sequences. Regions with dashed lines do not have at least one of the Cys and His amino acids and can no longer function as zinc-finger modules. Amino acids predicted to be damaging by SIFT are the white characters against black background; most occur in regions that no longer function as zinc-finger modules.