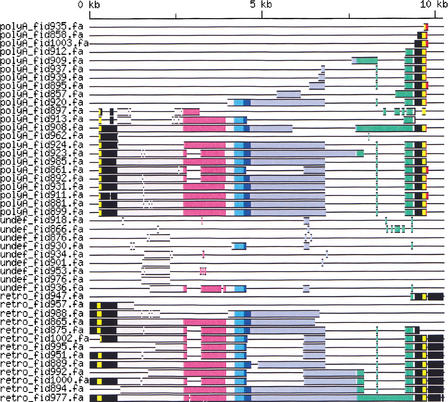
Figure 1.
Genomic structure of HERV-W copies on chromosomes 6 and 7 shown schematically as a multiple alignment of elements. Only internal sequences containing elements are included, 35 soloLTRs are not shown. Sequences were aligned to the Repbase Update consensus (see Methods). In the consensus of 3′ LTR, we introduced 50 A nucleotides to show the poly(A) tail (positions 9733–9782 in red). The LTR U5 and U3 regions are shown in black (positions 1–780 and 9407–10236), LTR R regions are in yellow. They were defined from TATA-box (TATAAA sequence, pos. 228–233 and 9634–9639 for the 5′ and 3′ LTR, respectively) to poly(A) signal (ATTAAA, pos. 308–313 and 9714–9719). ORFs are colored as follows: gag (pos. 2718–3923) in purple, pro (pos. 4192–4641) in cyan blue, pol (pos. 4450–7692) in light blue, and env (pos. 7720–9348) in green. The overlapping region of pro and pol is in dark blue. Other regions are contrasted in gray. In addition to several deletions, several splice variants are visible, see also Supplementary Figure 1S (online at http://www.genome.org). The last element, retro_fid977, is the syncytin-coding virus on chromosome 7.