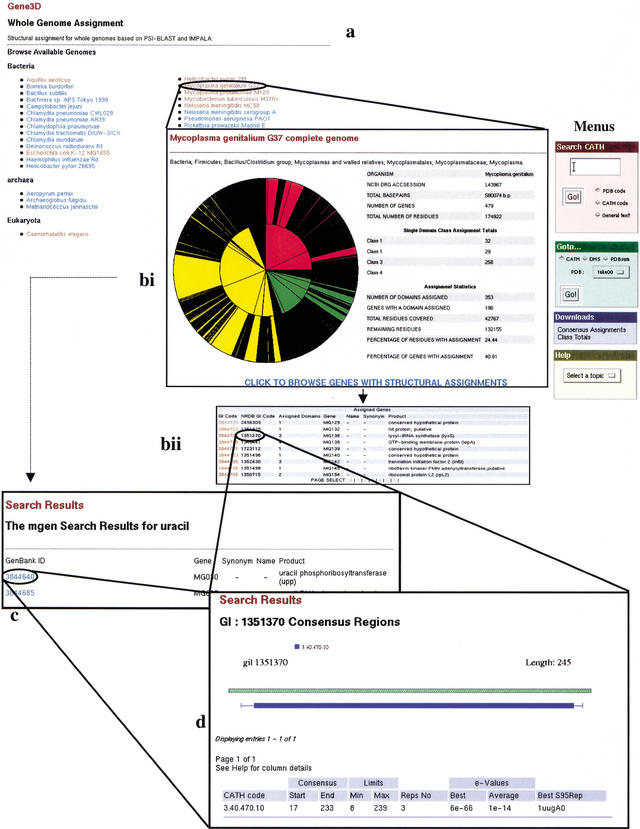
Figure 1.
An overview of the Gene3D server. (a) Genome Selection page. From here you can pick a genome to search. This brings up the assignment statistics page. Choosing ‘full’ also includes a summary of all the domains assigned to the genome. ‘Brief’ presents you with just the statistics. (b) Once a genome is selected, you get the statistics page. From here you can choose to search the genome using a keyword search; you can pick a gene from the list presented in the search results page (marked C). If you select a gene, you will go straight to that gene's domain assignments. (c) The Keyword search page. If you chose to search the genome using a key word (in this case, ‘uracil’), you will be presented with every gene in that genome which is associated with this key word. From here you can pick your gene of interest. (d) The assignment results page. If you chose a gene on the statistics page or on the search results page, you will be presented with the assignment results. These are presented as a diagram of the domain assignments (below the green hashed representation of the gene) and a summary of the PSI-BLAST results which led to this assignment (in the table below). From here you can link to the CATH database, the DHS, and PDBsum to gather further functional and structural information.