Figure 7.
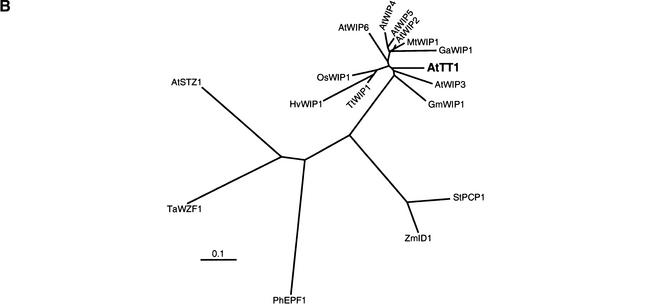
Relationship among various WIP proteins. (A) Amino acid sequence comparison of WIP domains. Part of the TT1 protein (Ler sequence) compared to the animal TFIIIA (Klug and Schwabe 1995) and the plant EPF (Takatsuji 1998) consensus sequences and aligned to partial TT1-like sequences. Only domains containing the sequence Trp-Ile-Pro (WIP, bold characters) are shown. Asterisks denote putative DNA binding residues. Black boxes indicate zinc binding residues. Conserved hydrophobic residues (h) are shaded. Amino acids conserved among all sequences are boxed. Dashes indicate gaps inserted to improve the alignment. Relative numbering without reference to a known start methionine is given in italics. (B) Distance analysis of several plant protein sequences containing two TFIIIA-type zinc finger motifs. For tree construction, only sequences of the zinc finger domains comparable to the WIP domain were used. See Table 1 for details on the factors included in the clustering. The scale bar indicates the distance in numbers of amino acid substitutions per site.