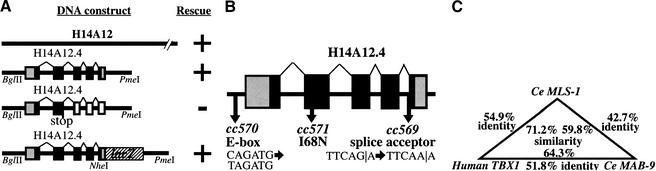
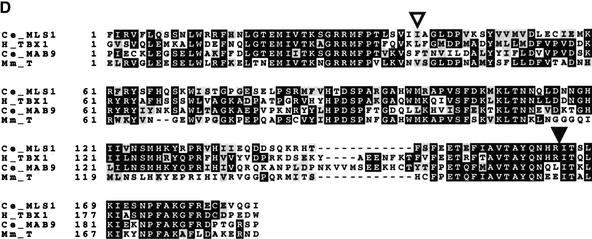
Figure 3.
Cloning mls-1. (A) (upper segment) A 4.4-kb BglII–PmeI subclone from fosmid H14A12 was able to rescue all mls-1 alleles. This fragment contains a single coding sequence as predicted by GeneFinder (P. Green, unpubl.). The T-box DNA-binding domain is colored black. (Middle segment) A clone containing an insertion at an AgeI site in the second exon causes a truncation at amino acid 109. The resulting sequence fails to rescue. (Lower segment) The structure of the mls-1::lacZ transgene is shown. LacZ was fused six amino acids from the C terminus of MLS-1. The construct results in the loss of the C-terminal six amino acids of MLS-1, but has the mls-1 3′ untranslated sequence. The mls-1::lacZ construct retains rescuing activity. (B) The mutations in mls-1 alleles cc569, cc570, and cc571 are shown. (C) Sequence comparison of CeMLS-1, human TBX1 (Chieffo et al. 1997), and CeMAB-9 (Woolard and Hodgkin 2000). Only residues within the T-box domain were compared. The genes have no similarity outside the T-box domain. Amino acid similarity was determined using the BLOSUM62 matrix (Henikoff and Henikoff 1992). (D) Sequence alignment of T-box domains. The alignment of the amino acid sequence of the T-box domain is shown for MLS-1 (Ce_MLS1), human TBX1 (H_TBX1; GenBank accession no. AF012130), C. elegans MAB-9 (Ce_MAB9; accession no. AJ252168), and mouse Brachyury (Mm_T; accession no. X51683). The start of the T-box domain of each protein is labeled 1. Identical amino acids are shown in black, similar amino acids are shaded. The MLS-1 sequence is based on GeneFinder prediction, homology-based sequence analysis, and a single allele defining the fourth splice acceptor. Positions of the mls-1 mutations in cc569 (closed arrowhead) and cc571 (open arrowhead) are shown.