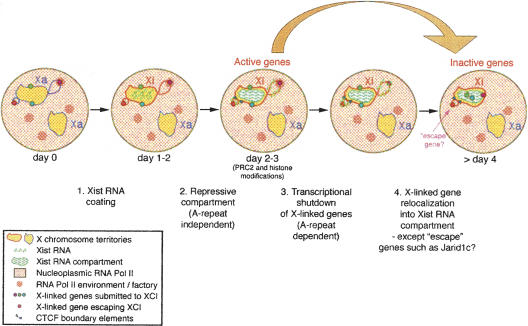
Figure 6.
Model for Xist RNA-mediated X-chromosome inactivation. In undifferentiated female ES cells, both X chromosomes are active (yellow). Only genes on the X chromosome undergoing inactivation (Xi) are depicted. (1) During early differentiation, Xist RNA starts to accumulate on the X chromosome chosen for inactivation. (2) The formation of this silent compartment is associated with the exclusion of the transcription machinery and the repression of internal repeat-rich regions. This step is independent of Xist A-repeats. (3) X-linked gene silencing is triggered via a Xist A-repeat-dependent mechanism, and histone modifications as well as PRC2/PRC1 accumulation (Xist A-repeat-independent) are observed. (4) Genes then become relocated into the Xist RNA compartment by default because they are no longer constrained by their requirement to be transcribed in an RNA Pol II-enriched environment (or potential transcription factory, see Osborne et al. 2004), and/or by an active mechanism of relocation, which may be dependent on the Xist A-repeats. This locus internalization may help maintain the silent state. Genes escaping inactivation, such as Jarid1c, have special elements (such as CTCF boundaries) that may prevent efficient internalization. Their location outside of the Xist repressive compartment could participate in, or facilitate, their reactivation.