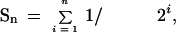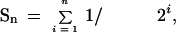SPIK gene disruption results in reduced pollen fitness. (
A) Histogram analysis of kanamycin-sensitive seedling percentage in the progeny of 31
SPIK/spik-1 plants (gray bars) and 44 control plants (white bars). The disrupting T-DNA (Fig.
1A) contains a gene (
nptII) conferring resistance to kanamycin. Sowing seeds from plants self-fertilized heterozygous (
SPIK/spik-1) or hemizygous for the transgene
nptII (control) on kanamycin medium (50 μg/mL) allowed the census (in percent) of homozygous wild-type seedlings (wt, free from disrupting T-DNA and thus kanamycin-sensitive) in the progeny of each plant. At least 300 seedlings were tested for each parental plant. Percentage data were sorted into eight classes for each parental genotype. The data (number of parent plants belonging to each class) were fitted by a Gaussian distribution. (
B) Evolution of the census of wild-type homozygous genotype through three subsequent self-fertilizing generations issued from
SPIK/spik-1 plants (gray symbols) or from control plants (white symbols). Thirty-five plants (F
0) of each genotype were grown. Their seeds (F
1) were harvested in bulk, of which at least 1200 seeds were sown on kanamycin medium (homozygous wt genotype census) and at least 100 seeds were sown on compost and grown in the greenhouse for producing the subsequent (F
2) generation. The same protocol was applied to analyze the F
2 seeds, and to produce and analyze the F
3 generation. It was assumed that pure self-fertilization occurred and that the
spik-1 mutation affected only pollen competitive ability. Within this framework, the predicted percentage of homozygous wild-type genotype in generation
n is %wt
n = 100Pwt × S
n, where
and Pwt is the probability of fertilization by wild-type pollen in competition with
spik-1 pollen. The value of the unknown parameter Pwt was adjusted to fit the data using a Marquardt–Levenberg algorithm (least-squares fitting). This yielded Pwt = 0.47 for the control genotype and Pwt = 0.64 for the
SPIK/spik-1 genotype.