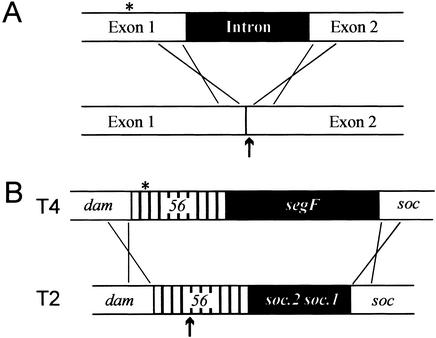
Figure 7.
Intron versus intronless homing. Filled boxes represent regions of DNA nonidentity. (A) Group I intron homing: An endonuclease encoded within the intron cleaves (↑) the intronless homolog close to the intron insertion site. DSB repair, which includes limited exonuclease digestion of cut DNA, incorporates the intron into the recipient, with frequency of coconversion of flanking markers (*) inversely proportional to their distance from the cleavage site. Insertion of the intron destroys the endonuclease recognition site. (B) Intronless homing by segF. SegF preferentially cleaves T2 gene 56, within a region of partial conservation with the T4 homolog (↑). Because of patchy conservation within gene 56, DSB repair initiates recombination in dam and soc, the closest regions of extensive sequence conservation. segF and the entire T4 gene 56 (with its nonpreferred SegF cleavage site) are transferred to recipients. The coconversion of markers in gene 56 (*) is close to 100%.