Figure 3.
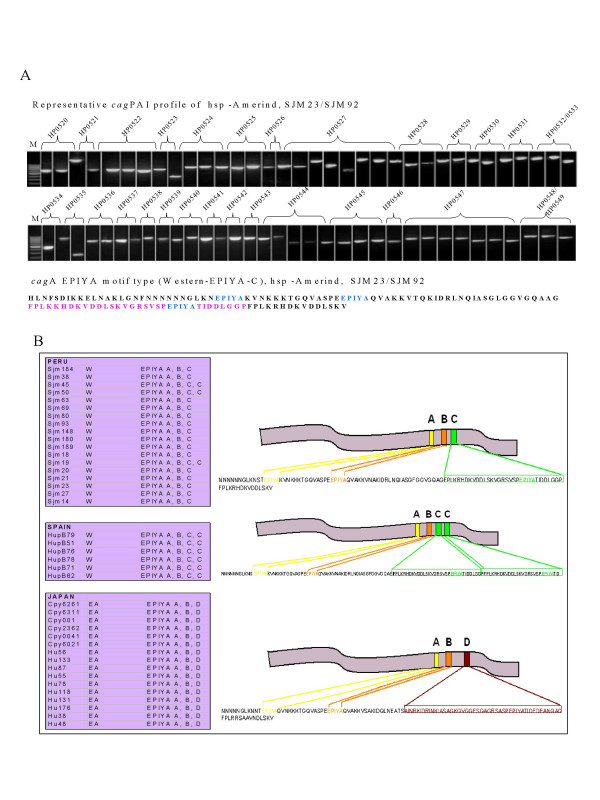
A. (Top) PCR based analysis of whole cagPAI of hsp-Amerind isolates from Peru. Overlapping primers spanning all of the constituent genes (see methods) amplified all the corresponding PCR products in the expected size range as described previously [30]. M indicates molecular weight marker (100 bp ladder). (Bottom) Amino acid signature of cagA (phosphorylation motifs – colored) – characteristic of modern cagA EPIYA (EPIYA C) as observed for all the SJM isolates from Peru. B. Pictorial depiction (right) of different types of cagA-EPIYA motif types prevalent in different H. pylori populations and their distribution in our isolates (boxes on the left).
