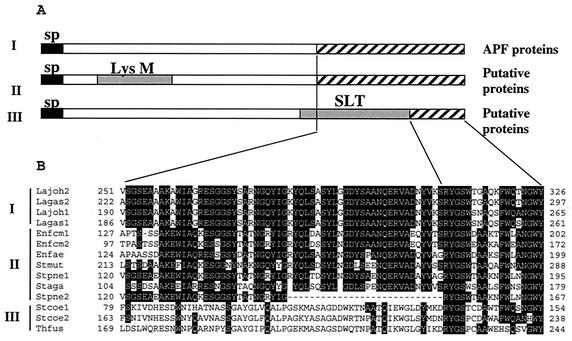
FIG. 3.
(A) Domain architecture of proteins containing conserved C termini. Functionally distinct proteins containing a conserved C terminus were clustered into three groups: group I, containing APF proteins, group II, containing putative proteins with a LysM domain (peptidoglycan binding motif), and group III, containing putative proteins with an SLT domain (lytic transglycosylases). Signal peptides (sp) are indicated by black boxes. Conserved domains LysM and SLT are in gray. The homologous sequence region is indicated by striped boxes. (B) Alignment of conserved C termini. CLUSTAL software was used (11). Lagas1, APF1 protein from L. gasseri 4B2; Lagas2, APF2 protein from L. gasseri 4B2; Lajoh1, APF1 protein from L. johnsonii NCC533 (La1) (accession no. AAN63951); Lajoh2, APF2 protein from L. johnsonii NCC533 (La1) (accession no. AAN63952); Enfcm1, a protein of Enterococcus faecium (accession no. NZ_AAAK01000149); Enfcm2, a protein of E. faecium (accession no. NZ_AAAK01000213); Enfae, LysM domain protein of Enterococcus faecalis (EF0443, The Institute for Genomic Research, unfinished genome); Stpne1, LysM domain protein of Streptococcus pneumoniae (accession no. NC_003028); Stpne2, a protein of S. pneumoniae R6 (accession no. NC_003098); Stmut, LysM domain protein from Streptococcus mutans (accession no. NP_722430); Staga, LysM domain protein from Streptococcus agalactiae (accession no. NC_004116); Stcoe1, a protein from Streptomyces coelicolor (accession no. AL357591); Stcoe2, a protein from S. coelicolor (accession no. AL391754); Thfus, a protein from Thermobifida fusca (DOE_2021, Joint Genome Institute, unfinished genome).