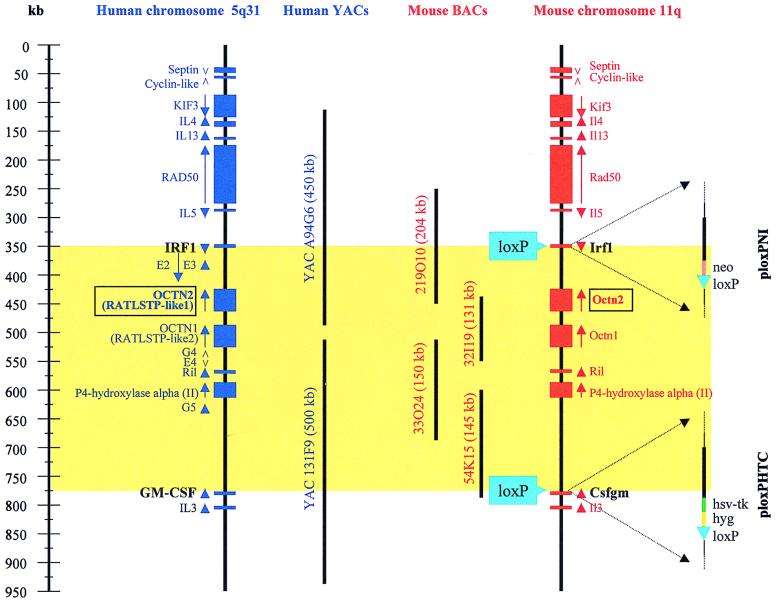
Figure 1.
The genomic organizations of the syntenic regions on mouse chromosome 11 and human chromosome 5q31. Computational analysis of human 5q31 sequences (9) identified putative genes by homology to known proteins (blue rectangles), by exact expressed sequence tag matches (E2, E3, E4, and E5) or by analysis of GRAIL-predicted exons (G4 and G5). For each gene, a vertical arrow indicates the direction of transcription. The genomic locations of the murine genes (red rectangles) in the diagram have been confirmed by content mapping of mouse chromosome 11 clones (YACs and BACs) and genomic DNA of homozygous deletion mice, as well as analyses of mouse chromosome 11 sequences (data not shown). The organization of the loxP targeting vectors, ploxPNI and ploxPHTC, introduced into the mouse genome and used to delete the 450-kb region between Irf-1 and Csfgm (shaded yellow) are shown on the right. Positions of the human YAC (medium-length bars) and mouse BAC (short bars) clones used to generate transgenic mice for the complementation studies are indicated. To create mouse BAC transgenics, BAC DNA was isolated by using an alkaline lysis method and was microinjected at a concentration of ≈1 ng/μl into fertilized FVB hybrid mouse eggs by using standard procedures. Transgenic mice were identified by PCR using primers that amplified vector sequences and genes encoded by the BAC sequences in transgenic homozygous deletion animals. Generation of the human YAC transgenic mice used in this study has been described previously (9).