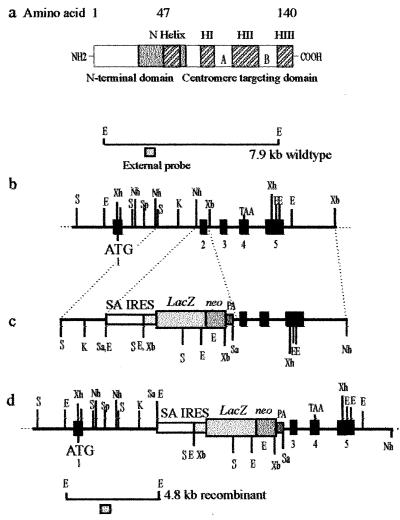
Figure 1.
Targeted disruption of the mouse Cenpa gene. (a) The mouse Cenpa protein showing the different subdomains, in particular those at the C terminus that are required for centromere targeting. Our targeting construct (see below) was designed to delete amino acids 29–64 (gray box), which will effectively remove the entire centromere-targeting domain. (b) A restriction map of the Cenpa gene. The exons are denoted by black boxes (23). (c) The gene replacement construct, where the selectable marker cassette consists of a splice-acceptor site (SA), a picornaviral IRES, a lacZ-neomycin-resistance fusion gene, and a simian virus 40 polyadenylation sequence (PA). (d) The Cenpa locus after gene disruption. The positions of external probes used in Southern analysis are shown and the expected size fragments are 7.9-kb wild-type allele and a 4.8-kb targeted allele. ATG and TAA are translation start and stop codons, respectively. Restriction enzymes used were SacI (S), SalI (Sa), EcoRI (E), XbaI (Xb), XhoI (Xh), KpnI (K), NheI (Nh), and SpeI (Sp).