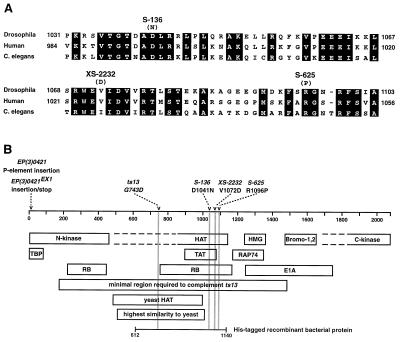
Figure 4.
Schematic representations of the identified mutations in TAF250. (A) Sequence comparison of a central region of Drosophila, human, and Caenorhabditis elegans with perfectly conserved amino acids highlighted (refs. 28 and 48; these sequence data were produced by the Sequencing Group at the Sanger Centre and can be obtained from ftp://ftp.sanger.ac.uk/pub). The C. elegans sequence is a conceptual translation of the genomic sequence so amino acid numbers are not available. The position and identities of amino acid changes in TAF250 alleles are indicated above the sequence. (B) A comparison of the sites of mutations in TAF250, indicated above, and functional domains indicated below a line depicting the TAF250 protein. In addition to mutations identified in this study, the position of the temperature-sensitive TAF250 mutation identified in the hamster cell line ts13 is indicated (49). The N- and C-terminal kinase domains (10), the HAT domain (11), two bromodomains (28), a high mobility group (HMG) box (28), and binding domains for TBP (18), HIV TAT (34), RAP74 (10), retinoblastoma (35), and adenovirus EIA (19) are indicated. Additional regions are indicated that are minimally required to complement the phenotypes caused by growth of ts13 cells at the nonpermissive temperature (50), display sequence similarity to the yeast TAF145 HAT domain (11), show the highest degree of sequence similarity to yeast TAF145 (51), and was used in the in vitro HAT assay.