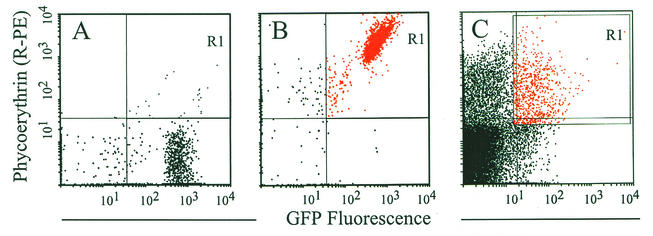
FIG. 4.
Labeling of bacteria in effusions improves FACS analysis. (A and B) GFP-expressing bacteria (strain 86-028NP/pKMM4B5) were incubated with a 1:50 dilution of a high-titer chinchilla antiserum directed against 86-028NP outer membrane proteins. Following incubation and successive washes, the bacterial pellet was resuspended in buffer without (A) or with (B) R-PE-labeled goat anti-human IgG (heavy plus light chains) at a concentration we had previously demonstrated to facilitate optimal cross-reactivity with the chinchilla antiserum (data not shown). GFP-expressing bacteria tagged with the PE-labeled antibody were detected in quadrant R1 (compare panels A and B). (C) Labeling of bacteria from effusions. This labeling methodology was used to identify GFP-expressing bacteria present in middle ear effusions, separating their signal from that of background, debris, and eukaryotic cells. Bacteria were separated from the effusions by setting the sort gate (R1) as shown.