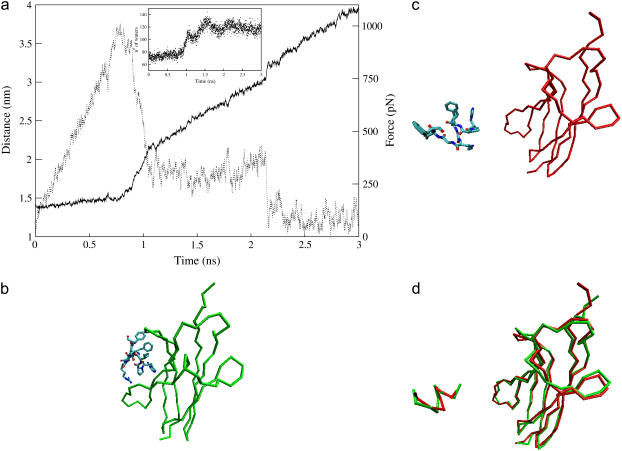
FIGURE 3.
Steered molecular dynamics of c2 moving the restraint point with a velocity of 0.5 nm/ns. (a) Distance between the mass centers of trkA-d5 and of the ACEHPIFGRGEFNME peptide (solid line) and force on the peptide (dashed line) plotted as a function of the simulation time. The peptide/receptor interactions are broken after 1.1 ns of SMD, except the salt bridges formed by the peptide residues Arg-9 and Glu-11, which break down at ∼1.45 ns and ∼2.15 ns of SMD, respectively. The inset shows the number of water oxygens within 0.45 nm of nonhydrogen atoms of the peptide residues, as a function of the simulation time. (b,c) Structure of c2 at the beginning (b) and at the end (c) of the simulation: the Cα trace of the receptor is represented (green at 5.7 ns of MD, red after 3 ns of SMD and further 5.4 ns of MD), along with the sticks representation of the peptide, colored by atom type. (d) Separated fit of the peptide and of the receptor structures in the initial (green) and the final (red) conformations (87).