TABLE 2.
TrkA-d5/N-term@NGF interactions
| NGF Residues | CS (nm2)
|
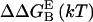 f.c. f.c. |
ΔΔ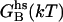
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1WWW | f.c. | c1 | c2 | 1WWW | f.c. | c1 | c2 | ||
| His-4 | 1.03 | 1.06 | 1.00 | 1.19 | 2.99 | 5.40 | 4.60 | 5.18 | 3.44 |
| Pro-5 | 0.47 | 0.51 | 0.57 | 0.36 | 1.47 | – | – | – | – |
| Ile-6 | 0.79 | 0.83 | 0.95 | 0.72 | −0.55 | 2.10 | 2.06 | 2.95 | 2.47 |
| Phe-7 | 0.39 | 0.49 | 0.54 | 0.25 | −0.84 | 1.27 | – | 1.44 | 1.39 |
| His-8(Gly) | 0 | 0 | 0 | 0 | 0 | – | 0.49 | – | – |
| Arg-9 | 0.44 | 0.57 | 0.53 | 1.06 | 2.52 | 0.29 | 2.11 | 2.06 | 2.99 |
| Gly-10 | 0.02 | 0.08 | 0.04 | 0.07 | – | – | – | – | – |
| Glu-11 | 0.65 | 0.68 | 0.90 | 0.83 | 6.33 | 0.69 | 2.50 | 1.83 | 2.82 |
| Phe-12 | 0.30 | 0.29 | 0.45 | 0.38 | −0.22 | 0.70 | 4.18 | 0.70 | 1.93 |
| Ser-13 | 0.34 | 0.39 | 0.10 | * | −2.13 | 0.35 | 0.35 | −0.22 | * |
Contact surface (CS) (61), electrostatic alanine scanning calculations (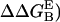 (65), and binding energy hot-spot predictions
(65), and binding energy hot-spot predictions 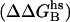 (48,49) are reported for NGF residues from His-4 to Ser-13. The NGF/trkA-d5 full-complex (f.c.) is the last MD snapshot of the previously performed simulation (47) and the c1 and c2 conformations are those after 5.7 ns of MD.
(48,49) are reported for NGF residues from His-4 to Ser-13. The NGF/trkA-d5 full-complex (f.c.) is the last MD snapshot of the previously performed simulation (47) and the c1 and c2 conformations are those after 5.7 ns of MD.
In c2, Ser-13 is absent.
