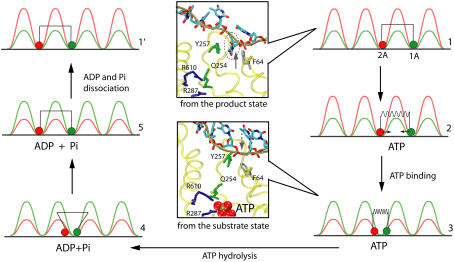
FIGURE 9.
Five-step PcrA translocation cycle. The figure shows schematically a translocation cycle for a mixed scenario (weak coupling as well as strong coupling; see text) involving configurations 1–5 as well as configuration 1′, which is equivalent to configuration 1, except moved forward by one nt. The mixed scenario involves both a loaded spring (nonzero potential Vσ′(x1, x2), transition 2→3) and a step with a random thermal motion (vanishing potential Vσ′(x1, x2), transition 4→5). In configurations 1, 2, and 1′, domain 1A (green) moves more readily than domain 2A (red), while it is the opposite for configurations 3–5. The inset figures show how the domain and ssDNA base motions are coupled to the chemistry at the ATP binding site: upon the approach of domains 2A and 1A, Arg-287, and Arg-610 move close to the γ-phosphate of ATP; Gln-254 is linked closely to Tyr-257, which forms a key binding pocket for an ssDNA base, but squeezes out the base when the domains approach each other in binding ATP; Gln-254 has been identified as a key participant in ATP hydrolysis along with the mentioned arginines (17). The suggested mechanism therefore involves three steps: 1), binding of ATP that pulls domain 1A toward domain 2A; 2), insertion of Arg-287 and Arg-610 into an optimal (for hydrolysis) position in the ATP binding pocket along with Gln-254 linked to a key ssDNA interaction site; and 3), rapid hydrolysis of ATP that initiates separation of domains 2A and 1A through movement of 2A alone.