Figure 6. G64S 3Dpol does not exhibit biased incorporation opposite ribavirin in the template.
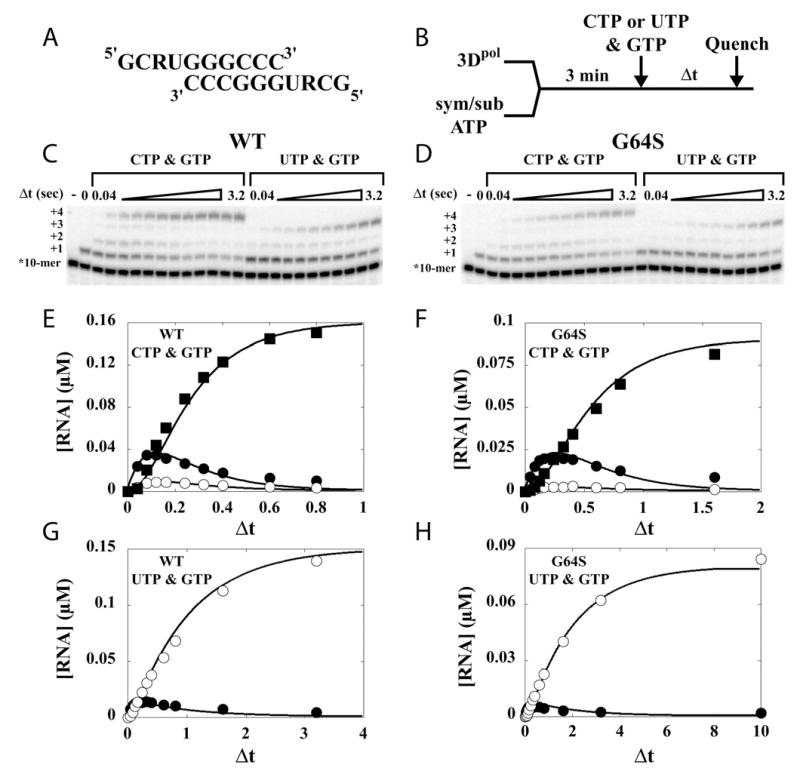
(A) Primer/template (sym/sub-UR) sequences. (B) Experimental design. 3Dpol (2 μM) was mixed with sym/sub (1 μM duplex) and ATP (5 μM) for 3 min to assemble 3Dpol-sym/sub product complexes after which CTP (1 mM) and GTP (500 μM) or UTP (1 mM) and GTP (500 μM) was added to the reaction. At fixed times after the addition of nucleotide the reaction was quenched by addition of EDTA. (C) Denaturing PAGE of the 32P-labeled products from WT 3Dpol-catalyzed nucleotide incorporation into sym/sub-UR. (D) Denaturing PAGE of the 32P-labeled products from G64S 3Dpol-catalyzed nucleotide incorporation into sym/sub-UR. (E) Kinetics of formation and disappearance of 12-mer (5 s−1), 13-mer (12 s−1) and 14-mer (50 s−1) by WT with CTP and GTP. (F) Kinetics of formation and disappearance of 12-mer (2.5 s−1), 13-mer (5 s−1) and 14-mer (50 s−1) by G64S with CTP and GTP. (G) Kinetics of formation and disappearance of 12-mer (1 s−1) and 13-mer (10 s−1) by WT with UTP and GTP. (H) Kinetics of formation and disappearance of 12-mer (0.5 s−1) and 13-mer (5 s−1) by G64S with UTP and GTP.
