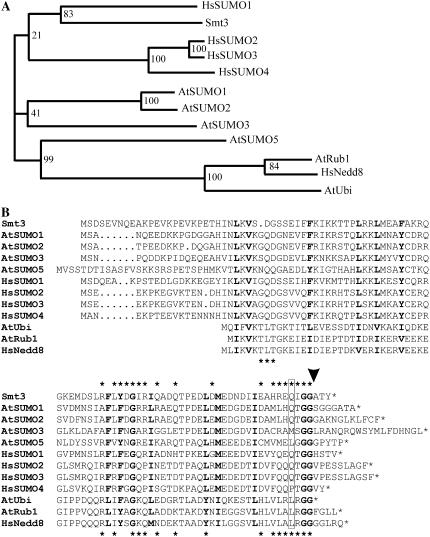
Figure 1.
Phylogenetic analysis and primary sequence alignment of SUMO and Nedd8 precursors from S. cerevisiae, Arabidopsis, and mammals. A, Proteins were grouped into a phylogram using the ClustalW server at the University of Wageningen (www.bioinformatics.nl/tools/clustalw.html) with standard settings. Bootstrap values (1,000 replicates) are shown at the branches. B, Amino acid residues conserved in all aligned sequences are depicted in bold letters. The C-terminal diglycine motif that is exposed during prepeptide processing is highlighted in bold letters, and the predicted cleavage site is marked by an arrowhead. A glutamine residue known to be important for the interaction of HsSUMO1 with the protease Senp2 is boxed. Asterisks depicted above and below the alignment indicate either Smt3 or HsNedd8 residues that are known to be in direct contact with the proteases ScULP1 or Den1, respectively.