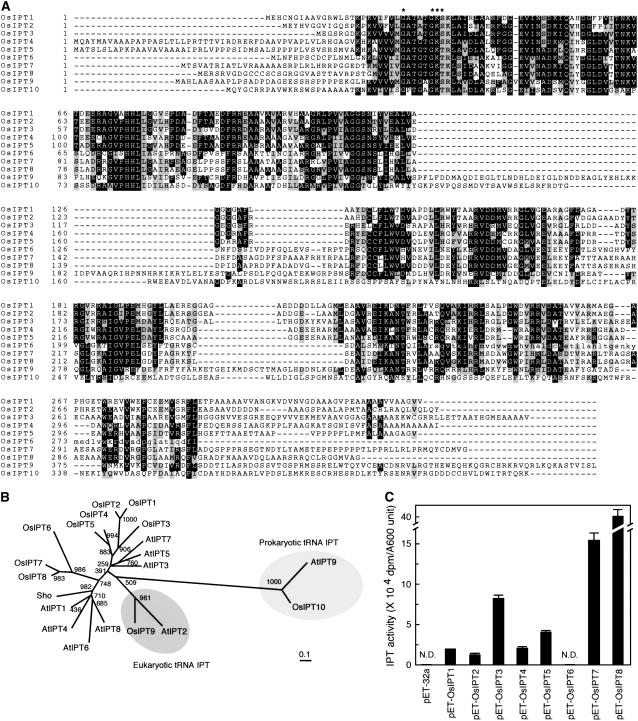
Figure 1.
Molecular and biochemical characterization of OsIPTs. A, Amino acid sequence alignment of OsIPTs. Exact matches are boxed in black; shaded boxes indicate conservative substitutions. Asterisks show conserved DMAPP-binding motifs. Lowercase letters indicate amino acid sequence of OsIPT6 from indica cultivar Kasalath. B, Unrooted dendrogram of IPT proteins in Arabidopsis (AtIPT1–AtIPT9), petunia (Sho), and rice (OsIPT1–OsIPT10). Bar, 0.1 amino acid substitutions per site. C, IPT activity of cell extracts. Crude extract of each transformant E. coli cell line was used to measure the IPT activity by radioisotope rapid assay. The amount of each sample for assay was equivalent to one A600 unit of cells. One A600 unit is defined as the amount of cells obtained from 1 mL of cell culture whose A600 value is 1.