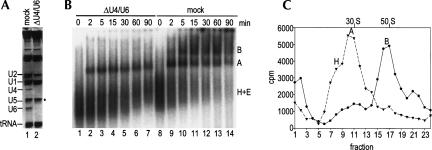
FIGURE 3.
U4/U6-depleted nuclear extract supports the formation of only spliceosomal A complexes. (A) RNA composition of U4/U6-depleted (lane 2) and mock-depleted (lane 1) extracts. RNA was analyzed as in Figure 2A. (B) Spliceosome assembly in U4/U6-depleted extract (lanes 1–7) and mock-depleted extract (lanes 8–14). Spliceosomal complexes were analyzed on a native gel at the indicated times. The positions of the H, A, and B complexes are indicated on the right. (C) Spliceosome assembly in mock- vs. U4/U6-depleted extract. Spliceosomal complexes were allowed to form under splicing conditions at 30°C for 20 min in U4/U6-depleted nuclear extract (triangles) and mock-depleted extract (circles). Subsequently, the complexes were separated on a 10%–30% glycerol gradient. The distribution of the radioactivity across each gradient is shown. Peaks were assigned as A or B complex based on their migration behavior according to Figure 1; the position of H complexes was verified independently (data not shown).