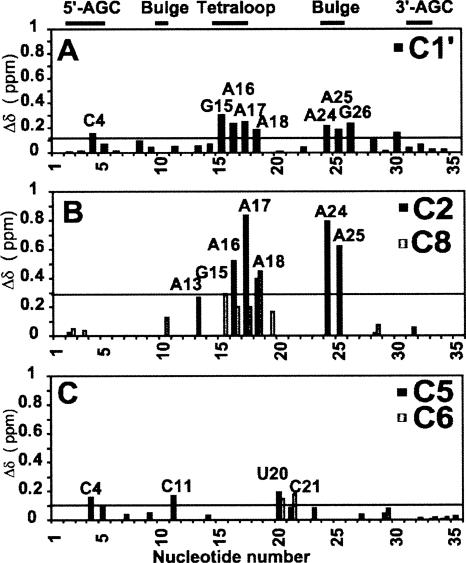
FIGURE 8.
Combined proton (1H) and carbon (13C) chemical shift perturbations {(Δδ) = [(Δ1H)2 + (Δ13Cχ)2]1/2} between free D5-PL and D123-bound D5-PL as a function of nucleotide sequence in (A) the ribose H1′–C1′ and aromatic (B) H2–C2 and H8–C8 and (C) H5–C5 and H6–C6 regions. Nucleotides that disappear at the titration end point (∼0.22 mM D123) are not shown. The horizontal line in each panel represents the calculated average chemical shift perturbation. χ = 0.18 and 0.24 for the base and the ribose carbons, respectively. The 3′-AGC label refers to nucleotides GUU that base-pair with the AGC triad nucleotides in the 5′-end of D5.