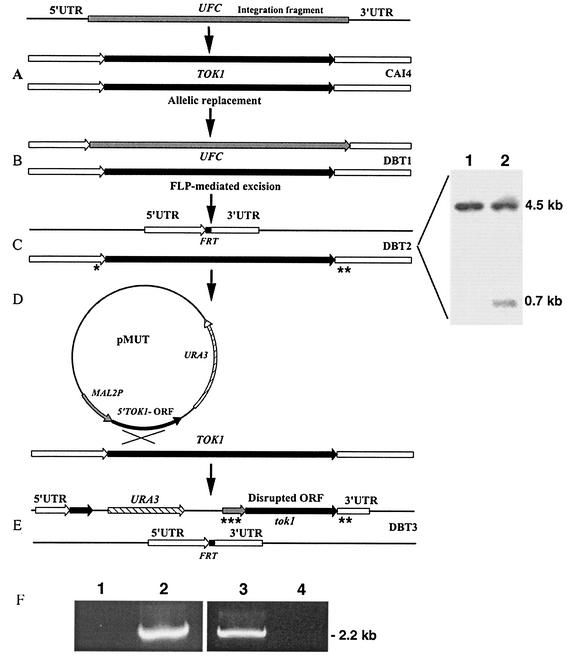
FIG. 1.
Construction of C. albicans TOK1-disrupted strains. Construction of C. albicans TOK1 knockout strains is described in Materials and Methods. (A) The integration fragment incorporating the UFC was introduced into the genome of the parental strain CAI4 (Ura−). (B) Integration of the UFC and replacement of the first TOK1 allele generates strain DBT1 (Ura+). (C) FLP-mediated excision of the UFC recycles the selectable marker and generates the single-allele deletion TOK1 strain DBT2 (Ura−). The small black box denotes the remaining single FRT site after UFC excision. Shown are the results of Southern hybridization analysis of BglII-digested genomic DNA from strains CAI4 (lane 1) and DBT2 (lane 2) with a 0.7-kb 5′ untranslated region (UTR) TOK1-specific probe. The sizes of the hybridizing fragments are as indicated: 4.5 kb, which corresponds to the intact TOK1 alleles in CAI4 (TOK1/TOK1) (lane 1), and 0.7 kb, which corresponds to the inactivated allele in strain DBT2 (TOK1/tok1) as well as the 4.5-kb fragment of the remaining wild-type allele (lane 2). (D) Integration of pMUT disrupts the second TOK1 allele. (E) Genomic structure of the null TOK1 strain DBT3 (Ura+). (F) PCR analysis of genomic DNA isolated from the homozygous (tok1/tok1) DBT3 (lanes 1 and 3) and the heterozygous (TOK1/tok1) DBT2 (lanes 2 and 4) strains. Lanes 1 and 2 were analyzed with the control primer combination TOK1Pctrl-f (*) and TOK1Uctrl-r (**) (Table 1), which amplify the remaining wild-type TOK1 allele of strain DBT2, while lanes 3 and 4 were analyzed with the primer combination MAL2Pctrl-f (***) and TOK1Uctrl-r (**) (Table 1), which amplify only the disrupted tok1 second allele. Asterisks on panels C and E indicate specific primer binding sites.